Похожие презентации:
Методы изучения регуляторных районов генов
1.
Методы изучения регуляторныхрайонов генов
Лекция I
Меркулова Татьяна Ивановна
Институт цитологии и генетики СО РАН
2.
Граничныйэлемент
MAR
Энхансер
Промоторный регуляторный район
Энхансер -4000
-40
+50
Базальный или коровый
промотор
-500
Ген
MAR
Граничный
элемент
Инициаторный комплекс
USA
CBP
Pol II
Mediator
IIE
IIB
IIA
IIF TAFs
TBP
IIH
Transcriptional Regulation in Eukaryotes: Concepts, Strategies, and Techniques. 2000, Cold Spring Harbor Laboratory Press
3.
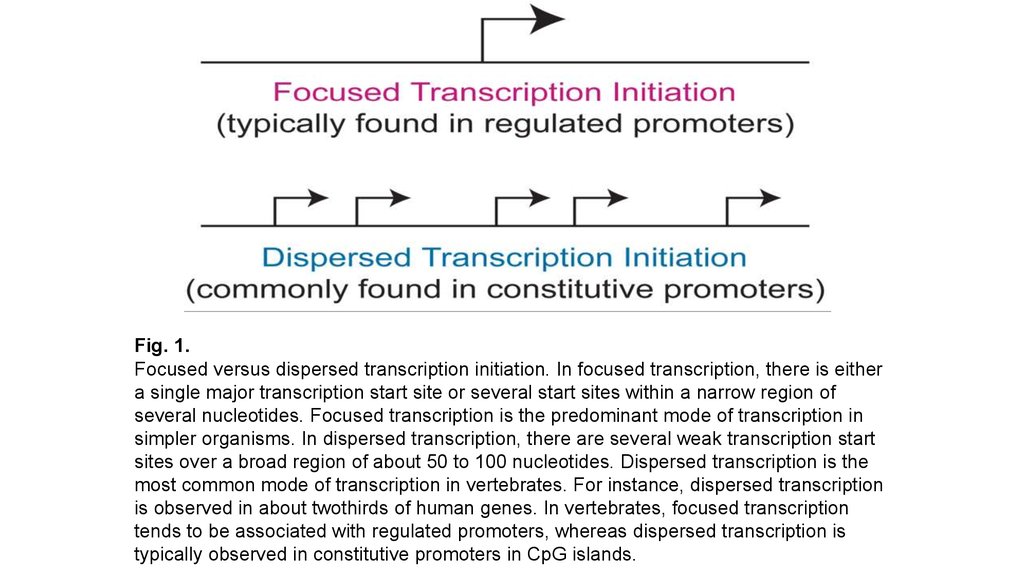
Fig. 1.Focused versus dispersed transcription initiation. In focused transcription, there is either
a single major transcription start site or several start sites within a narrow region of
several nucleotides. Focused transcription is the predominant mode of transcription in
simpler organisms. In dispersed transcription, there are several weak transcription start
sites over a broad region of about 50 to 100 nucleotides. Dispersed transcription is the
most common mode of transcription in vertebrates. For instance, dispersed transcription
is observed in about twothirds of human genes. In vertebrates, focused transcription
tends to be associated with regulated promoters, whereas dispersed transcription is
typically observed in constitutive promoters in CpG islands.
4.
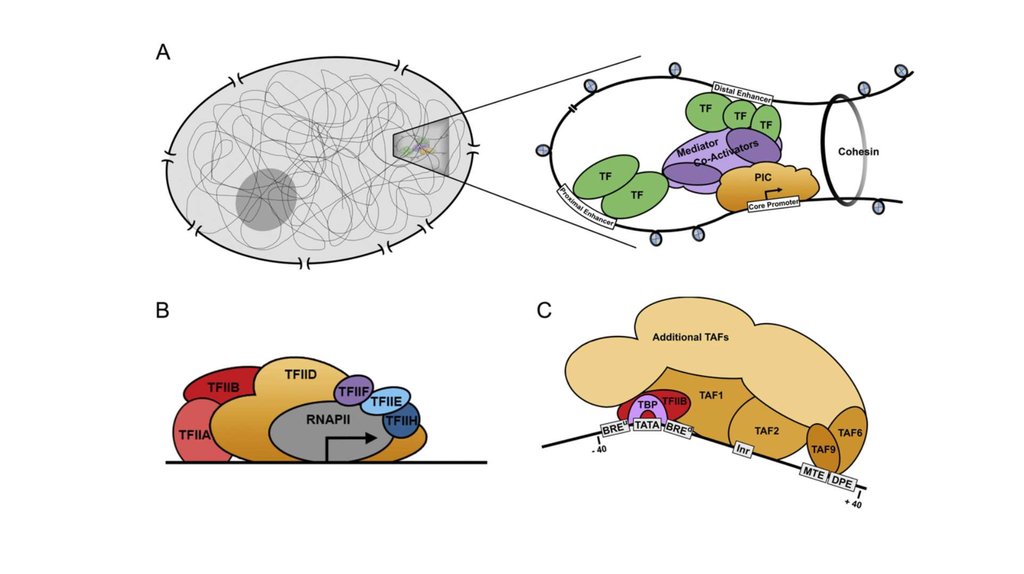
TFIID – комплекс TBF и ~10 TAFs-40 TFIIB
BRE
SSRCG
CC
TBP
TATA
TFIIB
BRE
TATAWAА RTDKK
R
K
TAF1 TAF2
TFIID
TAF6 TAF9
Inr
MTE
DPE
YYANWY
Y
CSARCSSAA
CGS
RGWYV
T
+40
5.
6.
7.
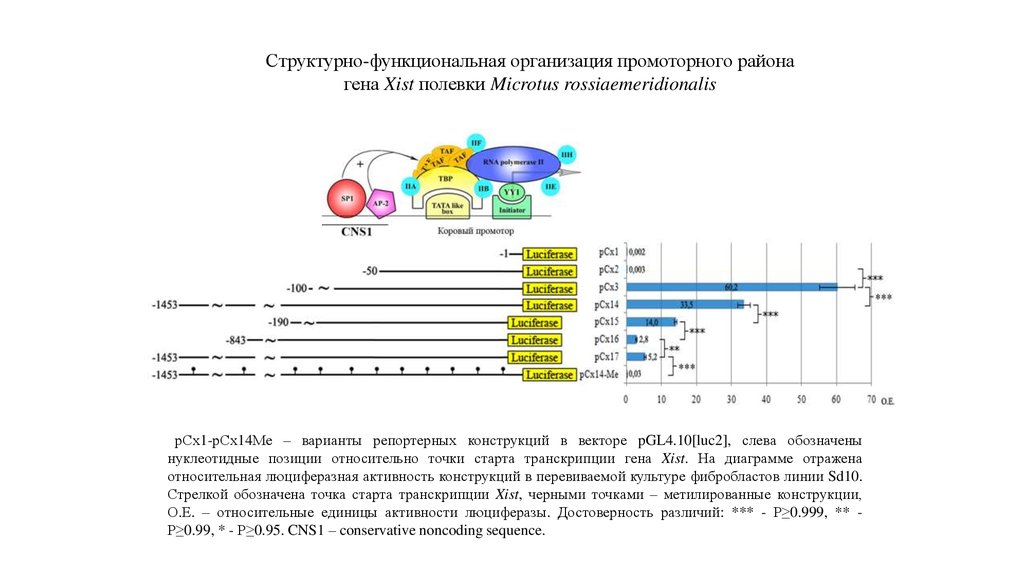
Структурно-функциональная организация промоторного районагена Xist полевки Microtus rossiaemeridionalis
рСх1-рСх14Ме – варианты репортерных конструкций в векторе pGL4.10[luc2], слева обозначены
нуклеотидные позиции относительно точки старта транскрипции гена Xist. На диаграмме отражена
относительная люциферазная активность конструкций в перевиваемой культуре фибробластов линии Sd10.
Стрелкой обозначена точка старта транскрипции Xist, черными точками – метилированные конструкции,
О.Е. – относительные единицы активности люциферазы. Достоверность различий: *** - P≥0.999, ** P≥0.99, * - P≥0.95. CNS1 – conservative noncoding sequence.
8.
9.
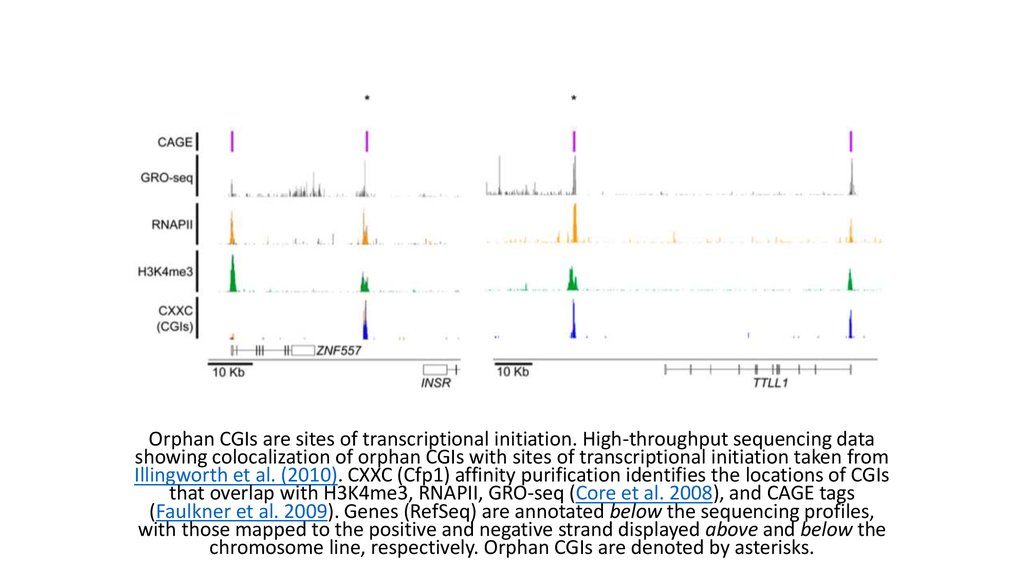
Orphan CGIs are sites of transcriptional initiation. High-throughput sequencing datashowing colocalization of orphan CGIs with sites of transcriptional initiation taken from
Illingworth et al. (2010). CXXC (Cfp1) affinity purification identifies the locations of CGIs
that overlap with H3K4me3, RNAPII, GRO-seq (Core et al. 2008), and CAGE tags
(Faulkner et al. 2009). Genes (RefSeq) are annotated below the sequencing profiles,
with those mapped to the positive and negative strand displayed above and below the
chromosome line, respectively. Orphan CGIs are denoted by asterisks.
10.
Diagram of the hGR Gene Structureand Organization
Untranslated exons 1A, 1B, or 1C are spliced to the same splice acceptor site in exon 2.
Promoters 1B and 1C are GC rich and are present in previously published sequences.
The hGR 1A promoter and exon are novel, and they are located approximately 27 kbp
upstream of the start site for transcription for exon 1C.
11.
Множественные экзоны 1Genomic organization of
mouse plectin (A), human
NOS1 (B), and rat GR (C)
genes.
Each gene contains a
tandem array of multiple first
exons in the variable region,
each of which is separately
spliced to a common set of
downstream constant
exons.The approximate
length of each gene is
shown below the
corresponding panels.
Theresa Zhang, Peter Haws and Qiang Wu. Multiple Variable First Exons: A Mechanism for Cell- andTissue-Specific Gene
Regulation. 2004 14: 79-89; Genome Res.
12.
Множественные экзоны 1Genome-wide distribution of human and mouse genes that have more than one first exon.
The numbers are shown above each histogram.The inset shows an enlargement of the
distribution of genes with more than three first exons.
Theresa Zhang, Peter Haws and Qiang Wu. Multiple Variable First Exons: A Mechanism for Cell- andTissue-Specific Gene
Regulation. 2004 14: 79-89; Genome Res.
13.
Синтез меченых на 5’ конце праймеровпри помощи γ- 32P и Т4 полинуклиотидкиназы
Гибридизация меченого праймера со
специфической мРНК
Удлинение праймера до 5’ конца мРНК
c помощью обратной транскриптазы и dNTP
Анализ полученной меченой ДНК
На секвенирующем геле
5’
3’
5’
3’
5’
3’
3’
5’
87nt
G
A T C
Extended
primer
87 nt
Размер меченого продукта соответствует
расстоянию 5’ конца праймера
до старта транскрипции
Free excess
primer
Transcriptional Regulation in Eukaryotes: Concepts, Strategies, and Techniques. 2000, Cold Spring Harbor Laboratory Press
14.
RACE ProcedureHybridize primer 100–200 nucleotides from 5′ end of mRNA.
Extend primer to 5′ end of mRNA using reverse transcriptase
and dNTPs.
Ligate oligo of known sequence to 3′ end of cDNA using RNA
ligase. Alternatively, extend cDNA using terminal transferase
(TdT) and dGTP.
Insert PCR
product into
vector
Sequence
Individual clones
Determine size on
sequencing gel (if
radiolabeled primer
used for PCR).
Perform PCR using primer complementary to ligated oligo and a
downstream primer that is slightly internal to the primer used for
cDNA synthesis above.
(Optional)
Sequence PCR products
Transcriptional Regulation in Eukaryotes: Concepts, Strategies, and Techniques. 2000, Cold Spring Harbor Laboratory Press
15.
Plasmid includes region of gene containingputativetranscription start site and 59 nucleotides
downstreamof this start site, fused to the SP6
promoter and flanked by convenient restriction sites.
RNase Protection
Cut with HindIII to linearize plasmid.
Add transcription buffer, SP6 RNA polymerase, NTPs plus [α -32P]UTP (asterisks) to generate
antisense probe.
Hybridize probe to isolated mRNA.
Digest with RNase T1 and RNase A
(cleaves single-stranded RNAs).
Creates a 59-nucleotide RNA-RNA hybrid.
Denature and analyze by denaturing gel electrophoresis.
Transcriptional Regulation in Eukaryotes: Concepts, Strategies, and Techniques. 2000, Cold Spring Harbor Laboratory Press
16.
Методы исследования регуляторных районовГетерологичный промотор
Интересующий промотор
Репортерный
ген
Репортерный ген
(Люцефераза или САТ)
ИЛИ
отдаленный
регуляторный район
Для анализа отдаленного
регуляторного района
Для анализа промотора
Трансфекция клеток
репортерной плазмидой
Измерение
активности
фермента
репортерного гена
Инкубация в течении 24-72 часов
транскрипция для эписомных плазмид и
синтез белка
Измерение уровня
репортерной мРНК
Transcriptional Regulation in Eukaryotes: Concepts, Strategies, and Techniques. 2000, Cold Spring Harbor Laboratory Press
17.
18.
Generation and initial analysis of more than 15,000 full-length humanand mouse cDNA sequences
Mammalian Gene Collection (MGC) Program Team*, 2000.
Robert L. Strausberg, Elise A. Feingold, Lynette H. Grouse, Jeffery G. Derge, Richard D. Klausner,
Francis S. Collins, Lukas Wagner, Carolyn M. Shenmen, Gregory D. Schuler, Stephen F. Altschul,
Barry Zeeberg, Kenneth H. Buetow, Carl F. Schaefer, Narayan K. Bhat, Ralph F. Hopkins,
Heather Jordan, Troy Moore, Steve I. Max, Jun Wang, Florence Hsieh, Luda Diatchenko, Kate Marusina,
Andrew A. Farmer, Gerald M. Rubin, Ling Hong, Mark Stapleton, M. Bento Soares, Maria F. Bonaldo,
Tom L. Casavant, Todd E. Scheetz, Michael J. Brownstein, Ted B. Usdin, Shiraki Toshiyuki,
Piero Carninci, Christa Prange, Sam S. Raha, Naomi A. Loquellano, Garrick J. Peters, Rick
D. Abramson, Sara J. Mullahy, Stephanie A. Bosak, Paul J. McEwan, Kevin J. McKernan, Joel A. Malek,
Preethi H. Gunaratne, Stephen Richards, Kim C. Worley, Sarah Hale, Angela M. Garcia, Laura J. Gay,
Stephen W. Hulyk, Debbie K. Villalon, Donna M. Muzny, Erica J. Sodergren, Xiuhua Lu, Richard
A. Gibbs, Jessica Fahey, Erin Helton, Mark Ketteman, Anuradha Madan, Stephanie Rodrigues,
Amy Sanchez, Michelle Whiting, Anup Madan, Alice C. Young, Yuriy Shevchenko, Gerard G. Bouffard,
Robert W. Blakesley, Jeffrey W. Touchman, Eric D. Green, Mark C. Dickson, Alex C. Rodriguez,
Jane Grimwood, Jeremy Schmutz, Richard M. Myers, Yaron S. N. Butterfield, Martin I. Krzywinski,
Ursula Skalska, Duane E. Smailus, Angelique Schnerch, Jacqueline E. Schein, Steven J. M. Jones, and
Marco A. Marra
Contributed by Francis S. Collins, et al.
19.
mRNAmRNA
mRNA
mRNA
mRNA
mRNA
mRNA
mRNA
mRNA
mRNA
mRNA
mRNA
20.
21.
The eukaryotic promoter database in its 30th year:focus on non-vertebrate organisms.
Dreos R., Ambrosini G., Groux R., Cavin Périer R., Bucher P.
Nucleic Acids Res. 2017 Jan 4;45(D1):D51-D55. doi: 10.1093/nar/gkw1069. Epub 2016 Nov 28.
22.
CAGE23.
24.
25.
26.
27.
28.
Карбокcи-терминальный домен самой большойсубъединицы полимеразы II содержит 52 повтора
гектапептида YSPTSPS
29.
30.
31.
The ENCODE Project ConsortiumENCODE Project Scientific Management:
National Human Genome Research Institute (E. A.
Feingold, P. J. Good, M. S. Guyer, S. Kamholz, L. Liefer,
K. Wetterstrand, F. S. Collins).
Initial ENCODE Pilot Phase Participants:
Affymetrix, Inc. (T. R. Gingeras, D. Kampa, E. A. Sekinger,
J. Cheng, H. Hirsch, S. Ghosh, Z. Zhu, S. Patel, A. Piccolboni,
A. Yang, H. Tammana, S. Bekiranov, P. Kapranov, R. Harrison,
G. Church, K. Struhl); Ludwig Institute for Cancer Research (B.
Ren, T. H. Kim, L. O. Barrera, C. Qu, S. Van Calcar, R. Luna, C.
K. Glass, M. G. Rosenfeld); Municipal Institute of Medical
Research (R. Guigo, S. E. Antonarakis, E. Birney, M. Brent, L.
Pachter, A. Reymond, E. T. Dermitzakis, C. Dewey, D. Keefe, F.
Denoeud, J. Lagarde, J. Ashurst, T. Hubbard, J. J. Wesselink,
R. Castelo, E. Eyras); Stanford University (R. M. Myers, A.
Sidow, S. Batzoglou, N. D. Trinklein, S. J. Hartman, S. F.
Aldred, E. Anton, D. I. Schroeder, S. S. Marticke, L. Nguyen,
J.Schmutz, J.Grimwood,M.Dickson, G. M. Cooper, E. A. Stone,
G. Asimenos, M. Brudno); University of Virginia (A. Dutta, N.
Karnani, C. M. Taylor, H. K. Kim, G. Robins); University of
Washington (G. Stamatoyannopoulos, J. A.
Stamatoyannopoulos, M. Dorschner, P. Sabo, M. Hawrylycz, R.
Humbert, J. Wallace, M. Yu, P. A. Navas, M. McArthur, W. S.
Noble); Wellcome Trust Sanger Institute (I. Dunham, C. M.
Koch, R.M.Andrews,G. K.Clelland, S. Wilcox, J. C. Fowler, K.D.
James, P. Groth, O. M. Dovey, P. D. Ellis, V. L. Wraight, A. J.
Mungall, P. Dhami, H. Fiegler, C. F. Langford, N. P. Carter,
D. Vetrie); Yale University (M. Snyder, G. Euskirchen, A. E.
Urban, U. Nagalakshmi, J. Rinn, G. Popescu, P. Bertone, S.
Hartman, J. Rozowsky, O. Emanuelsson, T. Royce, S. Chung, M.
Gerstein, Z. Lian, J. Lian, Y. Nakayama, S. Weissman, V. Stolc, W.
Tongprasit, H. Sethi).
Additional ENCODE Pilot Phase Participants:
British Columbia Cancer Agency Genome Sciences Centre (S. Jones,
M. Marra, H. Shin, J. Schein); Broad Institute (M. Clamp, K. LindbladToh, J. Chang, D. B. Jaffe, M. Kamal, E. S. Lander, T. S. Mikkelsen, J.
Vinson, M. C. Zody); Children’s Hospital Oakland Research Institute
(P. J. de Jong, K. Osoegawa,M.Nefedov, B. Zhu); National Human
Genome Research Institute/Computational Genomics Unit (A. D.
Baxevanis, T. G. Wolfsberg); National Human Genome Research
Institute/Molecular Genetics Section (F. S. Collins, G. E. Crawford, J.
Whittle, I. E. Holt, T. J. Vasicek, D. Zhou, S. Luo); NIH Intramural
Sequencing Center/National Human Genome Research Institute (E. D.
Green, G. G. Bouffard, E. H. Margulies, M. E. Portnoy, N. F. Hansen,
P. J. Thomas, J. C. McDowell, B. Maskeri, A. C. Young, J. R. Idol, R.
W. Blakesley); National Library of Medicine (G. Schuler); Pennsylvania
State University (W. Miller, R. Hardison, L. Elnitski, P. Shah); The
Institute for Genomic Research (S. L. Salzberg, M. Pertea, W. H.
Majoros); University of California, Santa Cruz (D. Haussler, D.
Thomas, K. R. Rosenbloom, H. Clawson, A. Siepel, W. J. Kent).
ENCODE Technology Development Phase Participants:
Boston University (Z. Weng, S. Jin, A. Halees, H. Burden, U. Karaoz,
Y. Fu, Y. Yu, C. Ding, C. R. Cantor); Massachusetts General Hospital
(R. E. Kingston, J. Dennis); NimbleGen Systems, Inc. (R. D. Green,M.
A. Singer, T. A. Richmond, J. E. Norton, P. J Farnham, M. J. Oberley,
D. R. Inman); NimbleGen Systems, Inc. (M. R. McCormick, H. Kim, C.
L. Middle, M. C. Pirrung); University of California,
et al!
The ENCODE Project Consortium. 2004. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306:
636–640.
32.
Functional genomic elements being identified by the ENCODE pilot phase.The indicated methods are being used to identify different types of
functional elements in the human genome
The ENCODE Project Consortium. 2004. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306:
636–640.
33.
Методы исследования регуляторных районовГетерологичный промотор
Интересующий промотор
Репортерный
ген
Репортерный ген
(Люцефераза или САТ)
ИЛИ
отдаленный
регуляторный район
Для анализа отдаленного
регуляторного района
Для анализа промотора
Трансфекция клеток
репортерной плазмидой
Измерение
активности
фермента
репортерного гена
Инкубация в течении 24-72 часов
транскрипция для эписомных плазмид и
синтез белка
Измерение уровня
репортерной мРНК
Transcriptional Regulation in Eukaryotes: Concepts, Strategies, and Techniques. 2000, Cold Spring Harbor Laboratory Press
34.
Comprehensive analysis of transcriptional promoter structure andfunction in 1% of the human genome
Sara J. Cooper, Nathan D. Trinklein, Elizabeth D. Anton, Loan Nguyen
and Richard M. Myers Genome Res., 2006 16: 1-10;
Clustergram of 642 putative promoter fragments.
The clustergram illustrates the hierarchical clustering of
promoter activity among 16 diverse cell lines.
Each row indicates the promoter activity of a fragment in
each of the cell lines, with red indicating the degree of
activity and black indicating no activity.
Promoter activity has been normalized and log
transformed to reflect comparable values between cell
lines.
Area A represents a cluster of promoter fragments with
strong, ubiquitous activity in all cell lines and area B
represents a cluster of promoter fragments that exhibit
variable function across the 16 cell types.
Comprehensive analysis of transcriptional promoter structure and function in 1% of the
human genome/ Sara J. Cooper, Nathan D. Trinklein, et al. Genome Res., 2006 16: 1-10


























 Биология
Биология








