Похожие презентации:
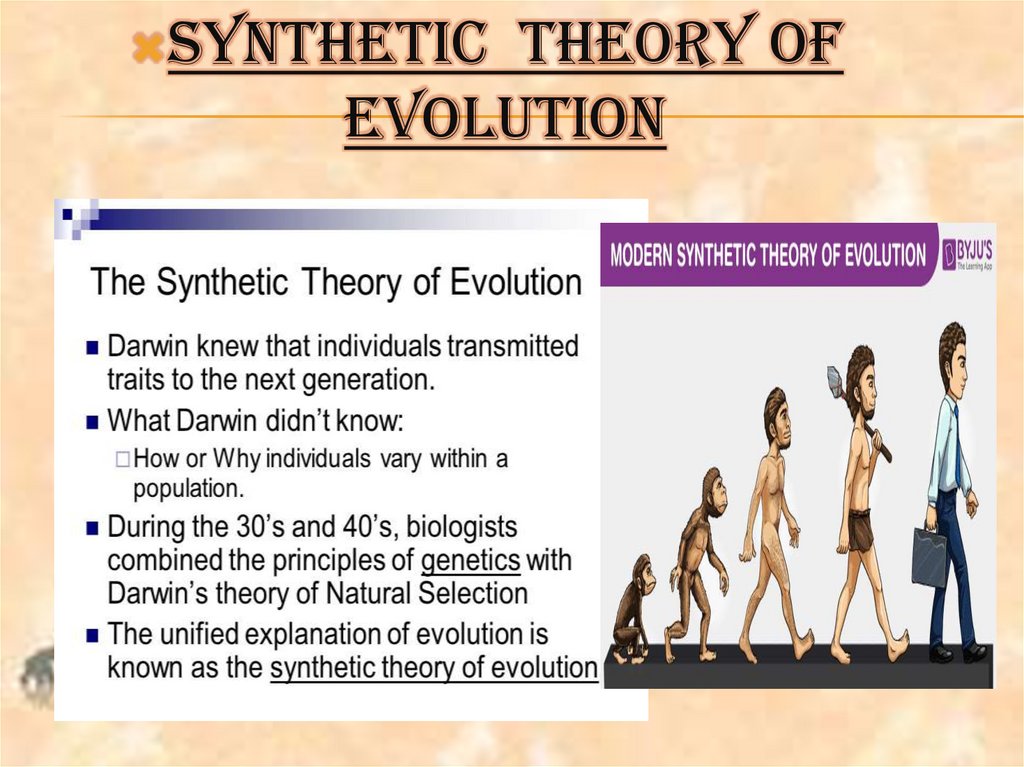
Synthetic theory of evolution
1.
DEPARTMENT OF BIOLOGYTOPIC:_SYNTHETIC THEORY OF EVOLUTION
GUIDED BY – SVETLANA SMIRNOVA
MADE BY-AISHWARAYA SHARMA
2.
SYNTHETICTHEORY OF
EVOLUTION
3.
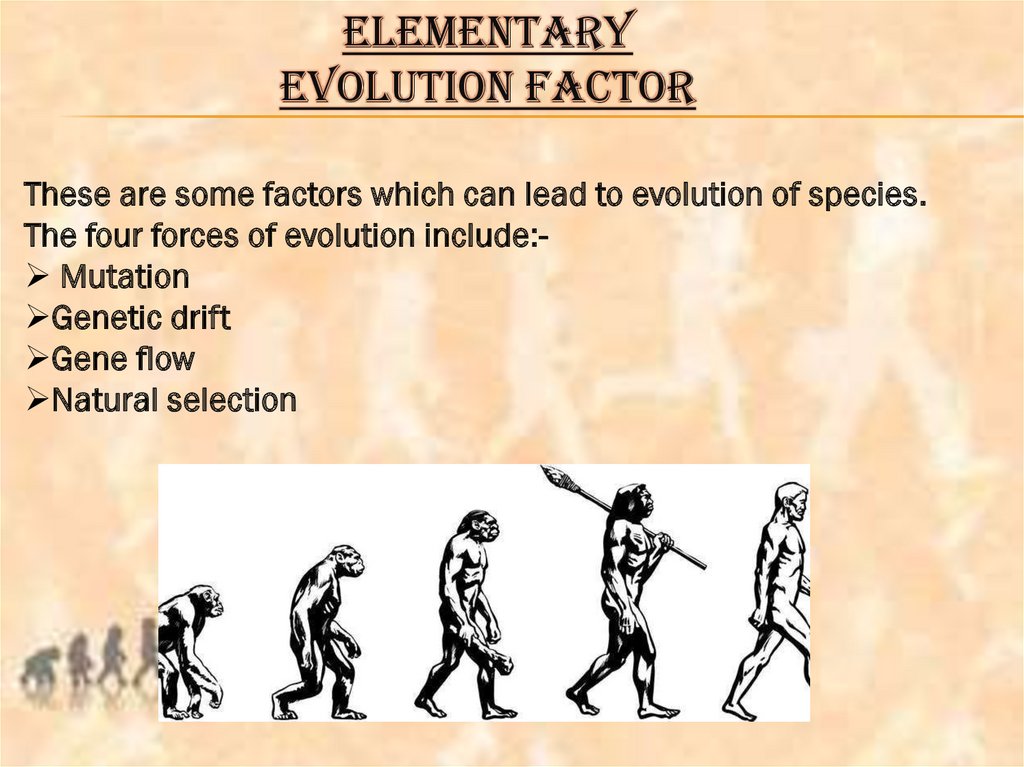
ELEMENTARYEVOLUTION FACTOR
These are some factors which can lead to evolution of species.
The four forces of evolution include: Mutation
Genetic drift
Gene flow
Natural selection
4.
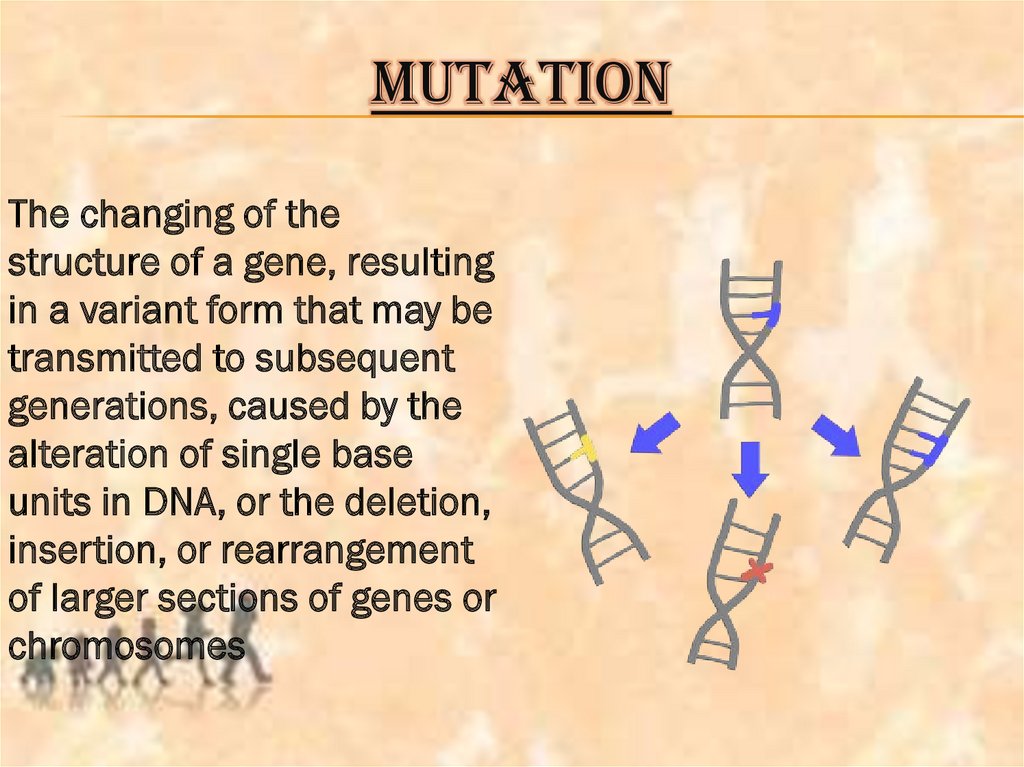
MUTATIONThe changing of the
structure of a gene, resulting
in a variant form that may be
transmitted to subsequent
generations, caused by the
alteration of single base
units in DNA, or the deletion,
insertion, or rearrangement
of larger sections of genes or
chromosomes
5.
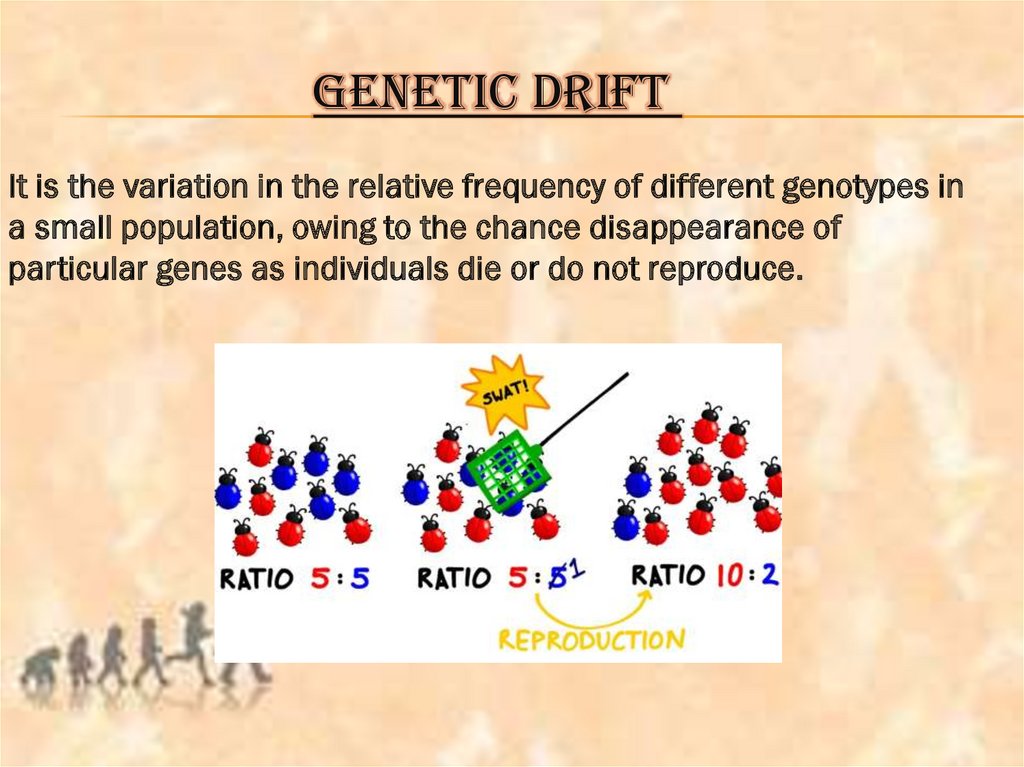
GENETIC DRIFTIt is the variation in the relative frequency of different genotypes in
a small population, owing to the chance disappearance of
particular genes as individuals die or do not reproduce.
6.
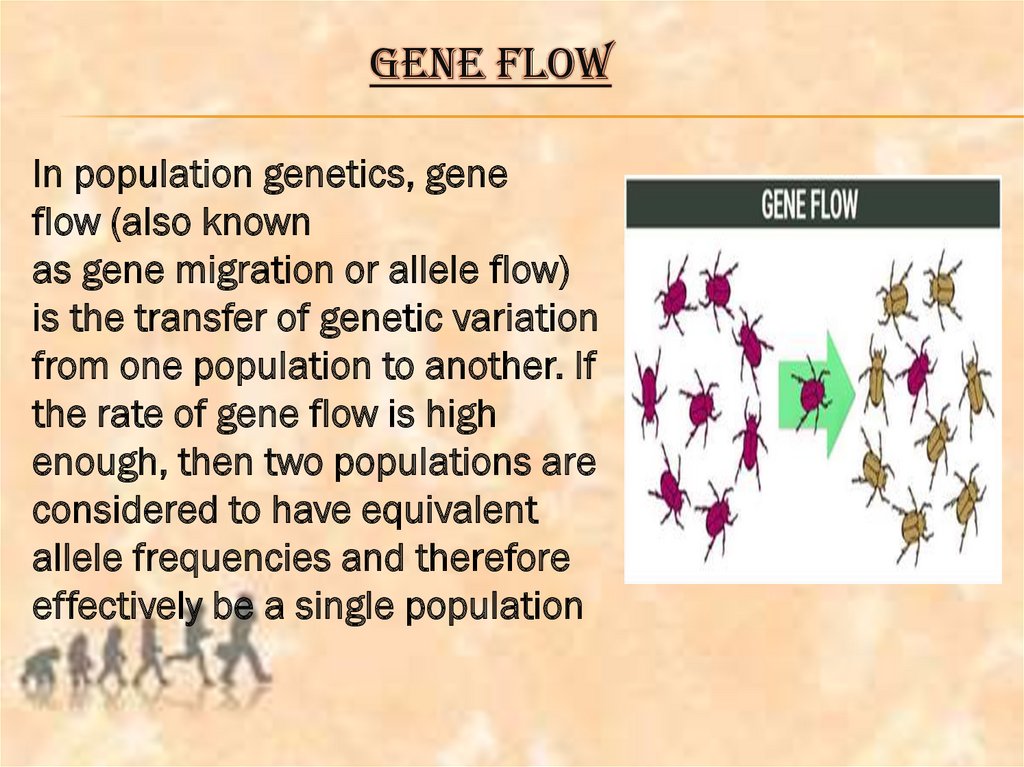
GENE FLOWIn population genetics, gene
flow (also known
as gene migration or allele flow)
is the transfer of genetic variation
from one population to another. If
the rate of gene flow is high
enough, then two populations are
considered to have equivalent
allele frequencies and therefore
effectively be a single population
7.
NATURAL SLECTIONThe process whereby organisms better
adapted to their environment tend to survive
and produce more offspring. The theory of its
action was first fully expounded by Charles
Darwin and is now believed to be the main
process that brings about evolution
8.
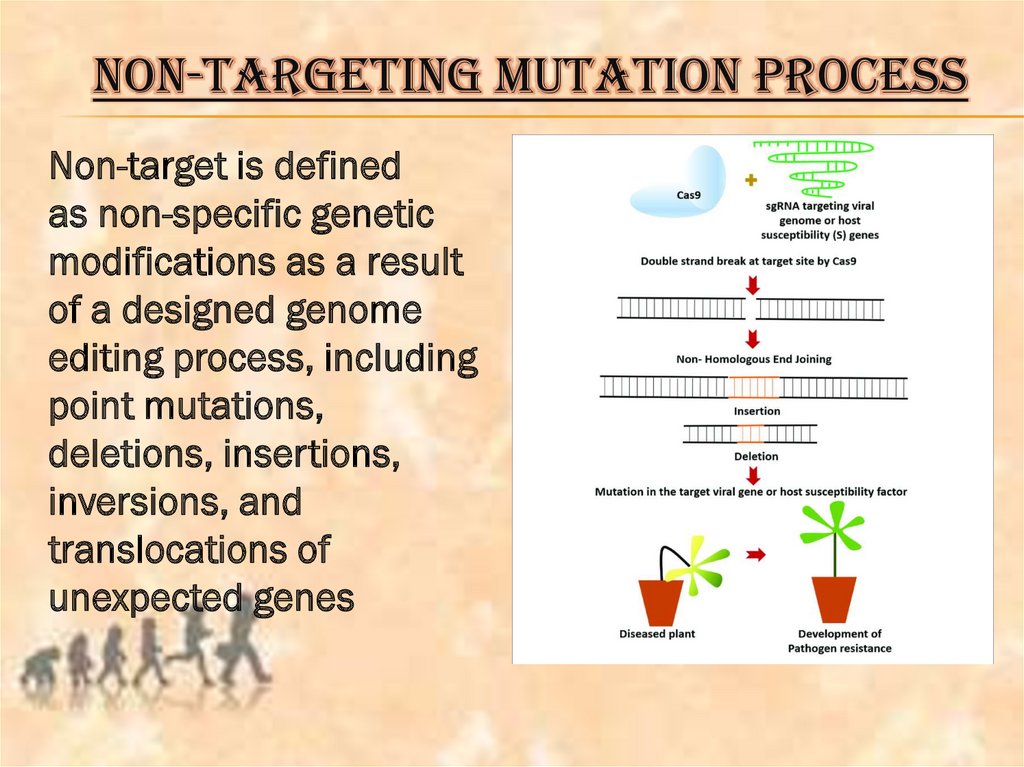
NON-TARGETING MUTATION PROCESSNon-target is defined
as non-specific genetic
modifications as a result
of a designed genome
editing process, including
point mutations,
deletions, insertions,
inversions, and
translocations of
unexpected genes
9.
MECHANISMThe CRISPR-Cas9 system works as the adaptive immune system in
bacteria and archaea.When a virus infects the bacteria, this system
incorporates segments of the viral DNA into the bacterial genome.
Upon a second invasion, transcripts from these sequences direct a
nuclease activity to its complementary sequence in the invading
virus so as to destroy it.
In order to extrapolate this method into eukaryotes in order to
develop a gene editing method, a Cas9 protein, a recognition
sequence RNA, and a transactivating RNA are required. The fusion
of both the recognition sequence specificity CRISPR RNA (crRNA)
and transactivating RNA (tracrRNA) is commonly used in
experiments and called a single guide RNA (sgRNA).[ It performs
both functions: the first 20 nucleotides of the sgRNA are
complementary to the DNA target sequence (cr function), while the
nucleotides following are part of a protospacer adjacent motif (PAM;
tracr function).[
10.
Off-targeting nuclease binding originates from a partial butsufficient match to the target sequence. Off-target binding
mechanisms can be grouped into two main forms: base
mismatch tolerance, and bulge mismatch.
11.
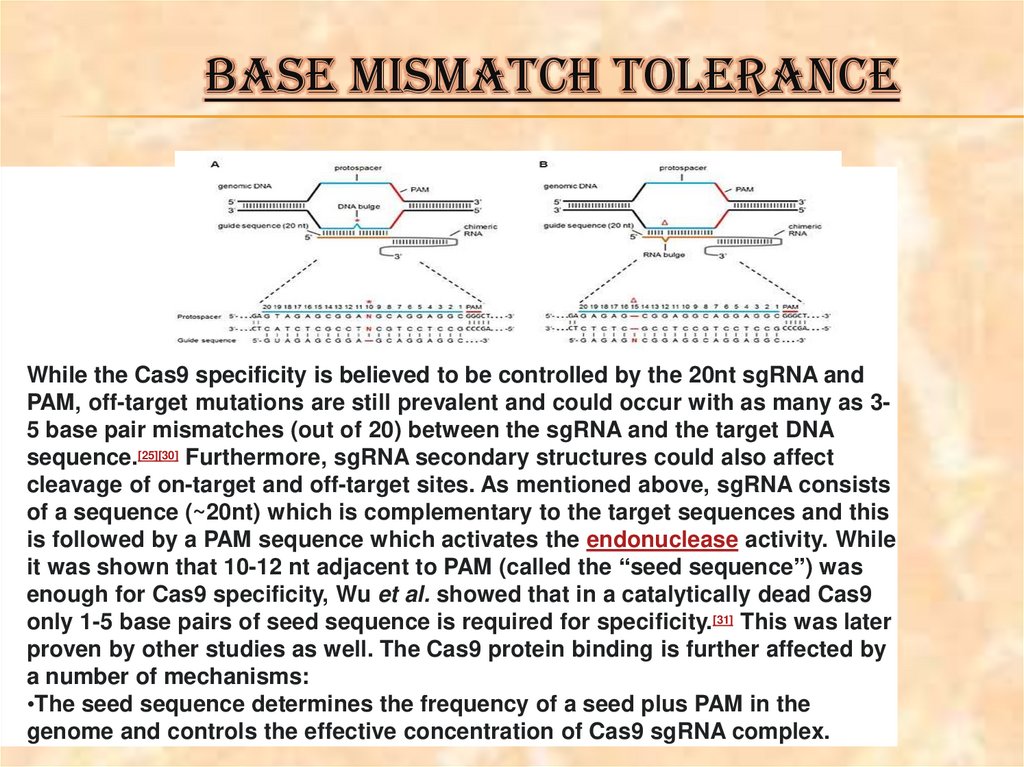
BASE MISMATCH TOLERANCEWhile the Cas9 specificity is believed to be controlled by the 20nt sgRNA and
PAM, off-target mutations are still prevalent and could occur with as many as 35 base pair mismatches (out of 20) between the sgRNA and the target DNA
sequence.[25][30] Furthermore, sgRNA secondary structures could also affect
cleavage of on-target and off-target sites. As mentioned above, sgRNA consists
of a sequence (~20nt) which is complementary to the target sequences and this
is followed by a PAM sequence which activates the endonuclease activity. While
it was shown that 10-12 nt adjacent to PAM (called the “seed sequence”) was
enough for Cas9 specificity, Wu et al. showed that in a catalytically dead Cas9
only 1-5 base pairs of seed sequence is required for specificity.[31] This was later
proven by other studies as well. The Cas9 protein binding is further affected by
a number of mechanisms:
•The seed sequence determines the frequency of a seed plus PAM in the
genome and controls the effective concentration of Cas9 sgRNA complex.
12.
Uracil-rich seeds are likely to have low sgRNA levels and increasespecificity since multiple uracil in the sequence can introduce
termination of the sgRNA transcription.[31][32]
Mismatches in the 5’ end of the crRNA are more tolerated as the
important site would be adjacent to the PAM matrix. Single and
double mismatches are also tolerated based on how to place it.
In a recent study, Ren et al. observed a link between mutagenesis
efficiency and GC content of sgRNA. At least 4-6bp adjacent to the
PAM are required for a good edit.[33]
While picking a gRNA, guanine is preferred over cytosine as the first
base of the seed adjacent to PAM, cytosine as the first in the 5’ and
adenine in the middle of the sequence. This design is based on
stability linked to formation of G quadruplexes.[31][32][34]
A ChIP was performed by Kim et al. showcasing that addition of a
purified Cas9 along with the sgRNA caused low off target effects
which means that there are more factors causing these effects. [35]
13.
VALUE OF MUTATION INEVOLUTION
Classical hypotheses of evolution have been derived by a comparison of the
morphological and other phenotypic properties of many different organisms.
The more information was accumulated and carefully evaluated, the fewer
hypotheses remained likely. More recently, evolution became explainable in
molecular terms, owing to the rapidly accumulating knowledge of the possible
alterations of DNA, the information transfer from DNA to RNA and proteins,
and the chemical and functional properties of proteins. This knowledge allows
one to state general rules about the evolution of macromolecules, rules that
hold for both simple and complex organisms. Some hypotheses of evolution
can thus be eliminated already by deduction from molecular principles rather
than by induction from many phenotypic observations. A complete theory of
evolution probably can be obtained only by a combination of both inductive
and deductive reasoning
14.
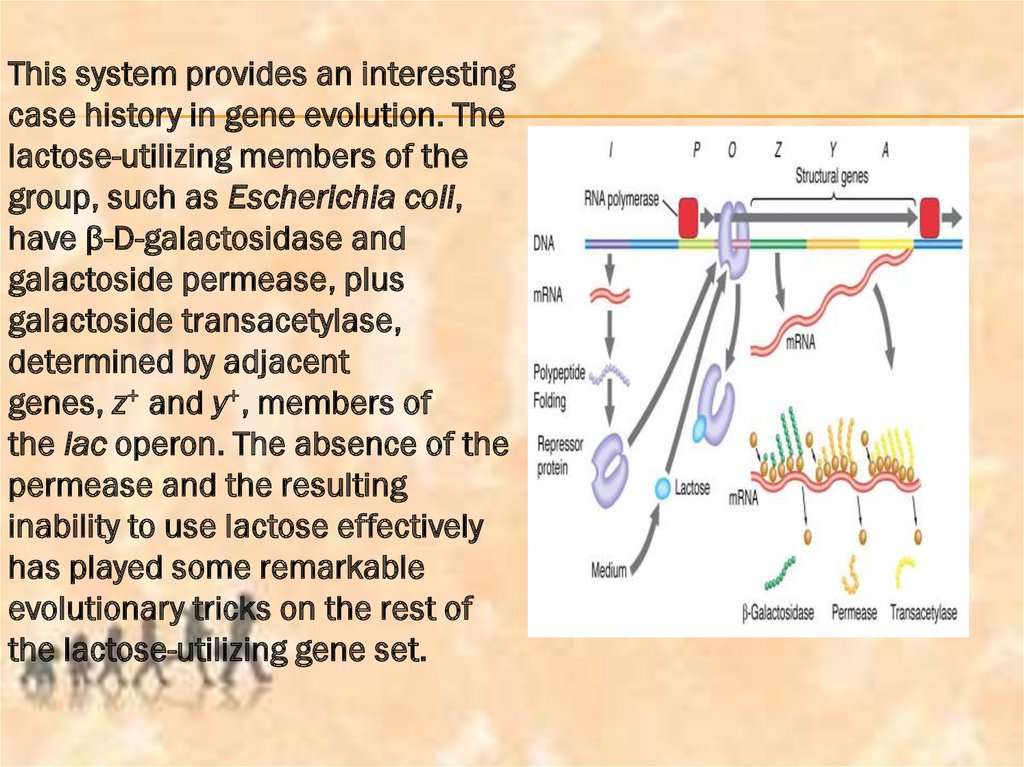
This system provides an interestingcase history in gene evolution. The
lactose-utilizing members of the
group, such as Escherichia coli,
have β-D-galactosidase and
galactoside permease, plus
galactoside transacetylase,
determined by adjacent
genes, z+ and y+, members of
the lac operon. The absence of the
permease and the resulting
inability to use lactose effectively
has played some remarkable
evolutionary tricks on the rest of
the lactose-utilizing gene set.
15.
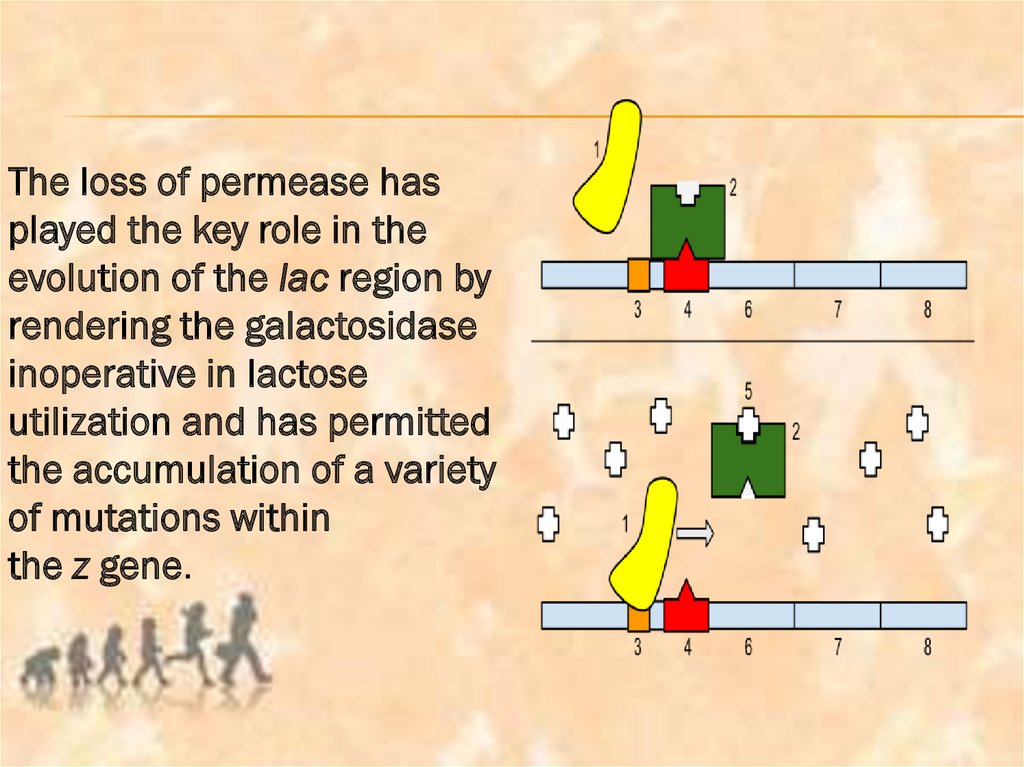
The loss of permease hasplayed the key role in the
evolution of the lac region by
rendering the galactosidase
inoperative in lactose
utilization and has permitted
the accumulation of a variety
of mutations within
the z gene.
16.
Proteins and genes might haverequired the simultaneous evolution
of the cell. Studies of the
organismic evolution of proteins
began with considerations of
applying sequential and terminal
residue methods to tracing
evolution of primary Sructure of
proteins.
17.
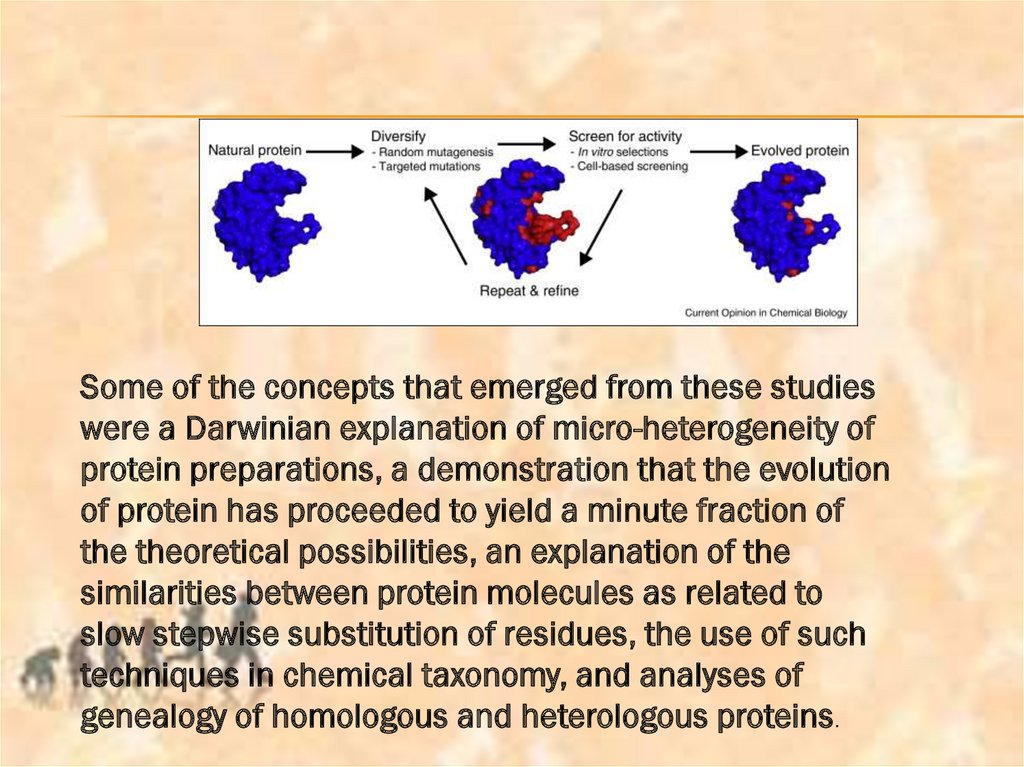
Some of the concepts that emerged from these studieswere a Darwinian explanation of micro-heterogeneity of
protein preparations, a demonstration that the evolution
of protein has proceeded to yield a minute fraction of
the theoretical possibilities, an explanation of the
similarities between protein molecules as related to
slow stepwise substitution of residues, the use of such
techniques in chemical taxonomy, and analyses of
genealogy of homologous and heterologous proteins.
18.
EFFECTS OF MUTATIONSince all cells in our body contain DNA, there are lots of places for mutations to
occur; however, some mutations cannot be passed on to offspring and do not
matter for evolution. Somatic mutations occur in non-reproductive cells and won't
be passed onto offspring. For example, the golden color on half of this Red
Delicious apple was caused by a somatic mutation. Its seeds will not carry the
mutation.The only mutations that matter to large-scale evolution are those that
can be passed on to offspring. These occur in reproductive cells like eggs and
sperm and are called germ line mutations.
19.
EFFECTS OF Germline MUTATION1.No change occurs in phenotype.
Some mutations don't have any noticeable effect
on the phenotype of an organism. This can
happen in many situations: perhaps the mutation
occurs in a stretch of DNA with no function, or
perhaps the mutation occurs in a protein-coding
region, but ends up not affecting the amino
acid sequence of the protein.
2.Small change occurs in phenotype.
A single mutation caused this cat's ears to curl
backwards slightly.
3.Big change occurs in phenotype.
Some really important phenotypic changes, like
DDT resistance in insects are sometimes caused
by single mutations. A single mutation can also
have strong negative effects for the organism.
Mutations that cause the death of an organism are
called lethals — and it doesn't get more negative than that.







 Биология
Биология








