Похожие презентации:
IMPRS workshop Comparative Genomics
1.
IMPRS workshopComparative Genomics
18th-21st of February 2013
Lecture 4
Positive selection
2.
What is positive selection?3.
Positive selection is selection on a particular trait- and the increased frequency of an allele in a population
4.
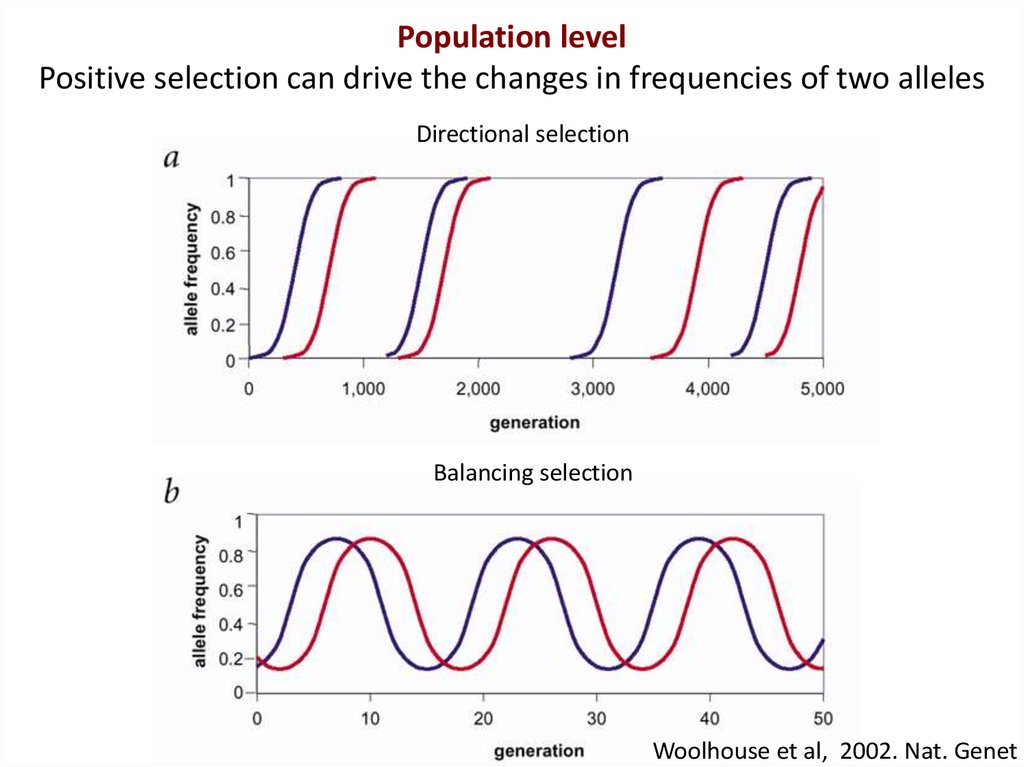
Population levelPositive selection can drive the changes in frequencies of two alleles
Directional selection
Balancing selection
Woolhouse et al, 2002. Nat. Genet
5.
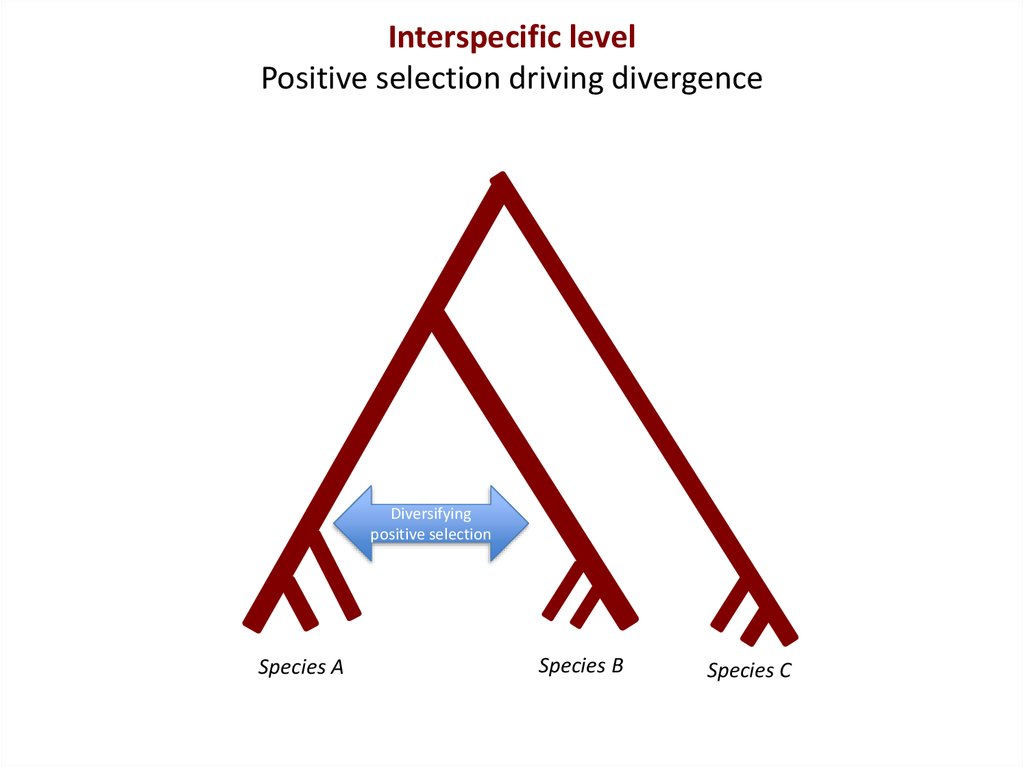
Interspecific levelPositive selection driving divergence
Diversifying
positive selection
Species A
Species B
Species C
6.
Why is it interesting to identify traits whichhave undergone or are under positive selection?
Function
Evolution
Environment
……
7.
How can we detect positive selection?8.
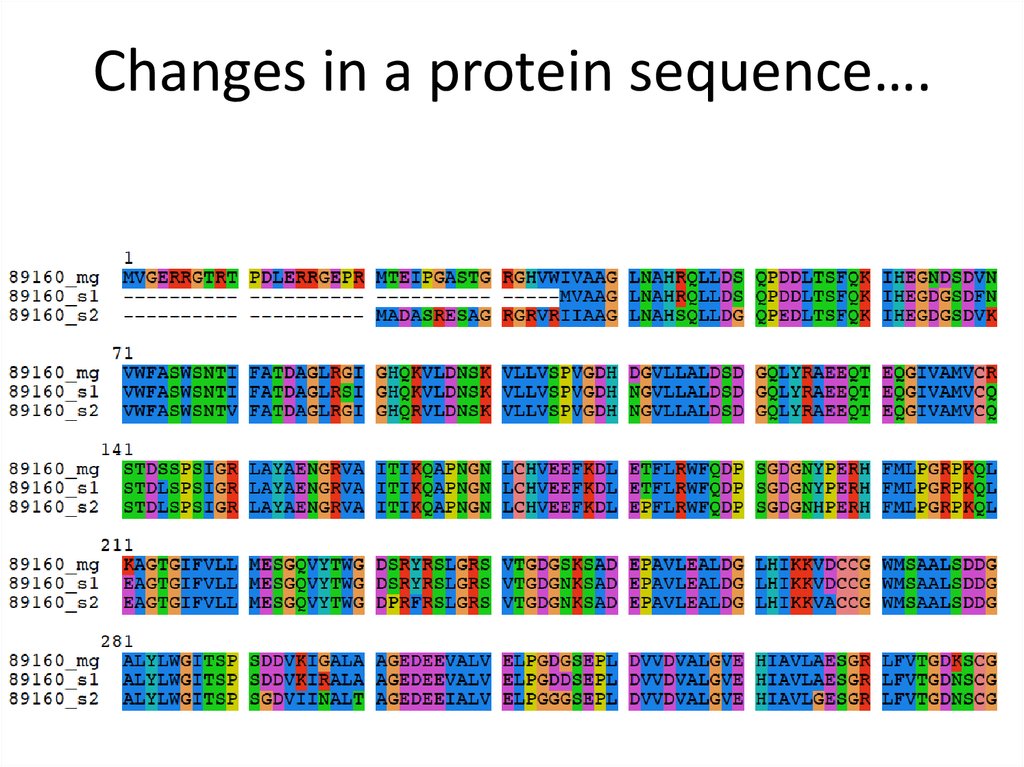
Changes in a protein sequence….9.
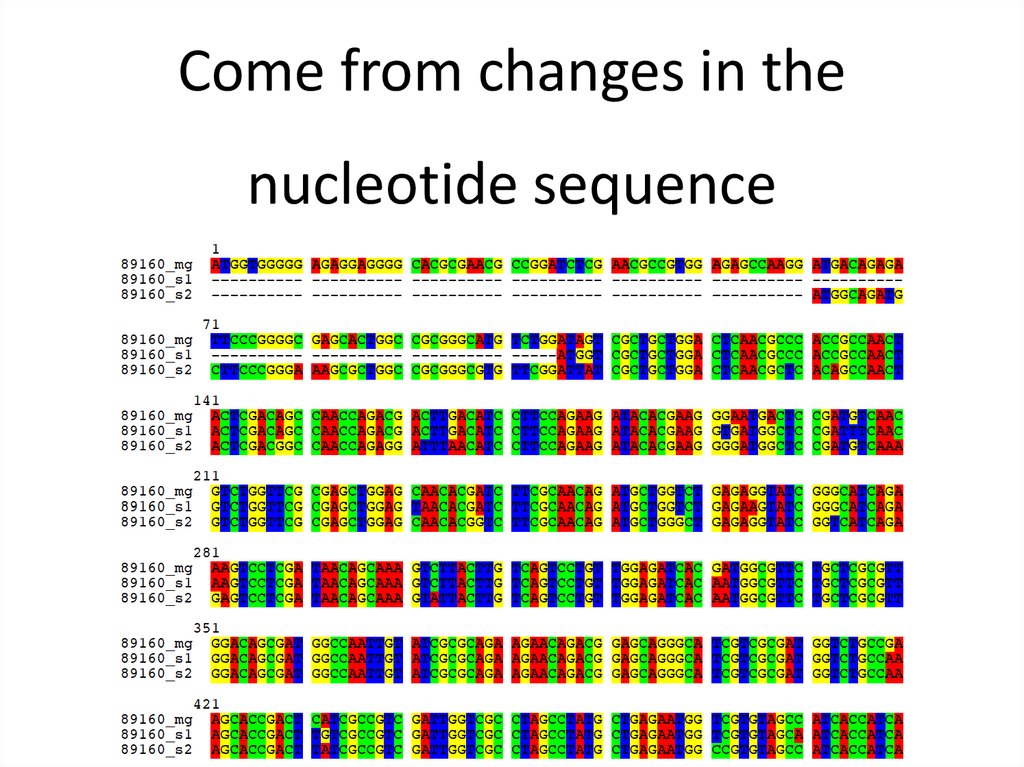
Come from changes in thenucleotide sequence
10.
Quantifying non-synonymous variation- an estimate of positive selection
Synonymous mutations: neutral mutations
Non-synonymous mutations: non-neutral mutations
11.
To measure positive selection:Rate of synonymous mutations
Rate of non-synonymous mutations
12.
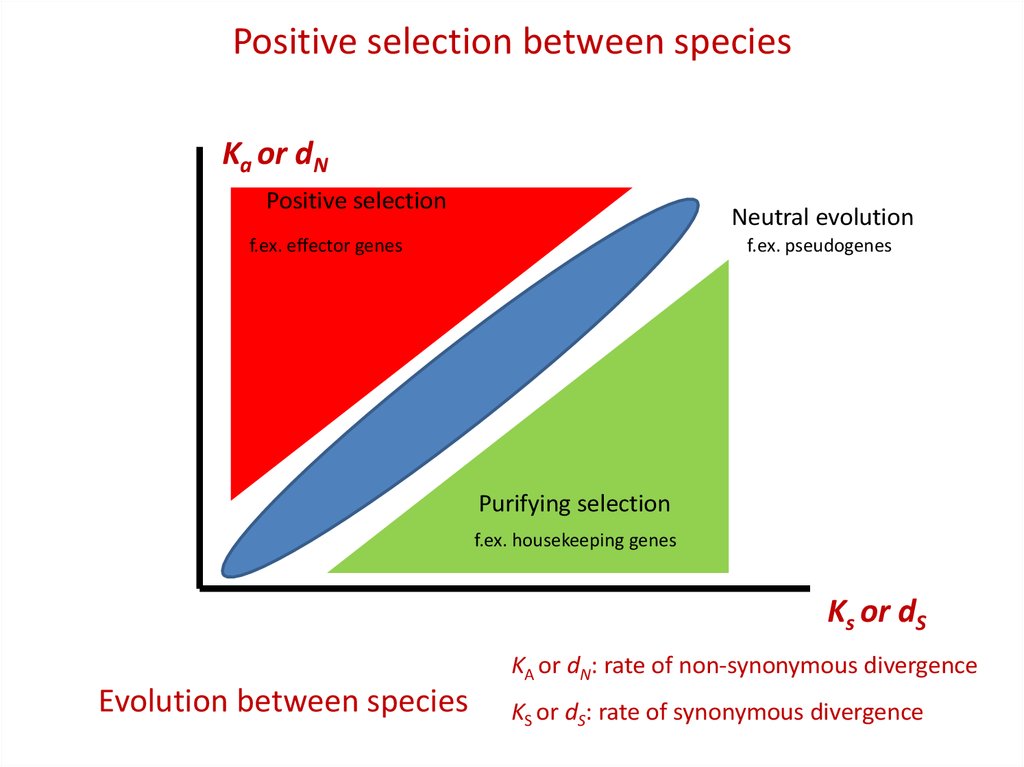
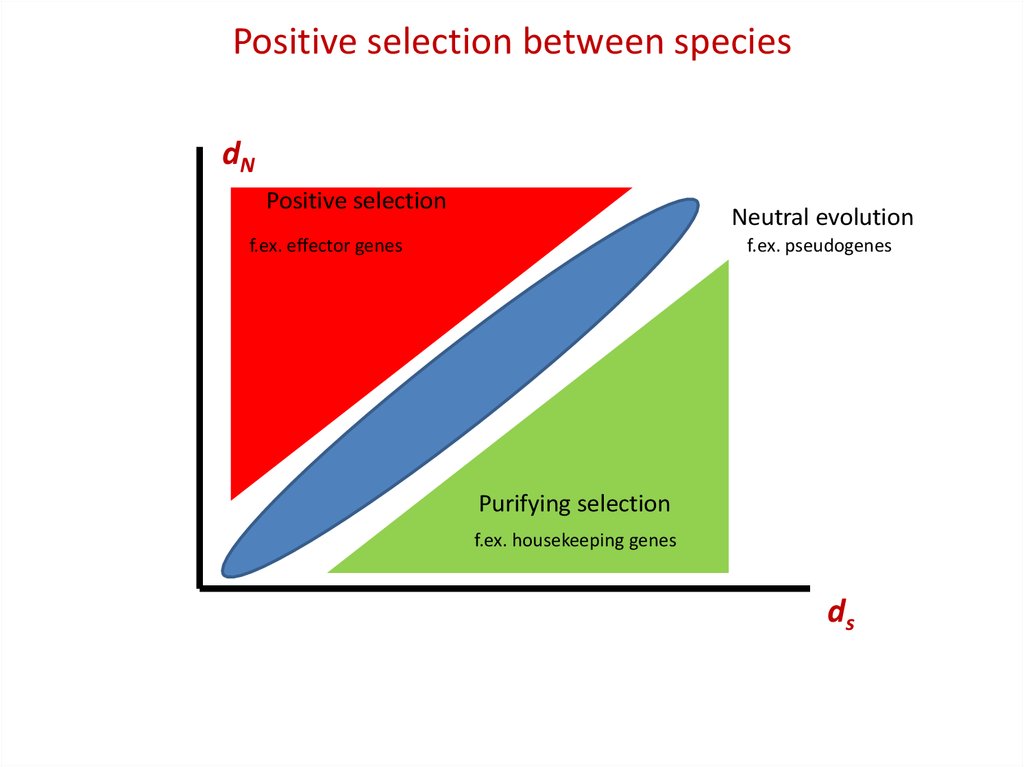
Positive selection between speciesKa or dN
Positive selection
Neutral evolution
f.ex. effector genes
f.ex. pseudogenes
Purifying selection
f.ex. housekeeping genes
Ks or dS
KA or dN: rate of non-synonymous divergence
Evolution between species
KS or dS: rate of synonymous divergence
13.
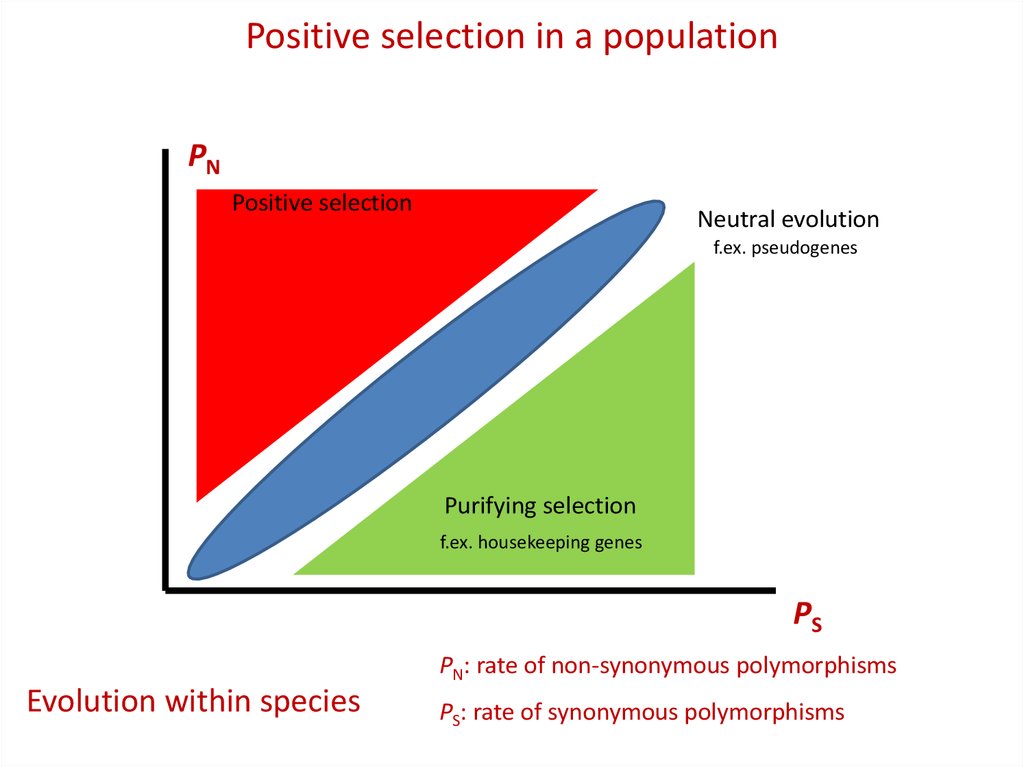
Positive selection in a populationPN
Positive selection
Neutral evolution
f.ex. pseudogenes
Purifying selection
f.ex. housekeeping genes
PS
PN: rate of non-synonymous polymorphisms
Evolution within species
PS: rate of synonymous polymorphisms
14.
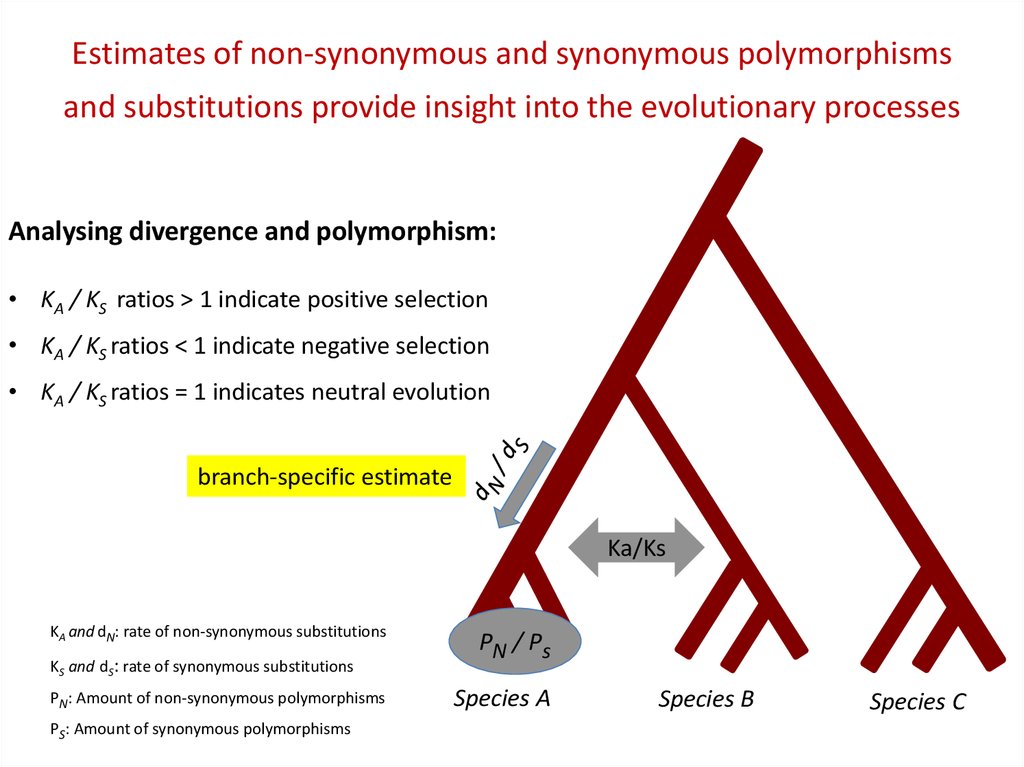
Estimates of non-synonymous and synonymous polymorphismsand substitutions provide insight into the evolutionary processes
Analysing divergence and polymorphism:
• KA / KS ratios > 1 indicate positive selection
• KA / KS ratios < 1 indicate negative selection
• KA / KS ratios = 1 indicates neutral evolution
branch-specific estimate
Ka/Ks
KA and dN: rate of non-synonymous substitutions
KS and dS: rate of synonymous substitutions
PN: Amount of non-synonymous polymorphisms
PS: Amount of synonymous polymorphisms
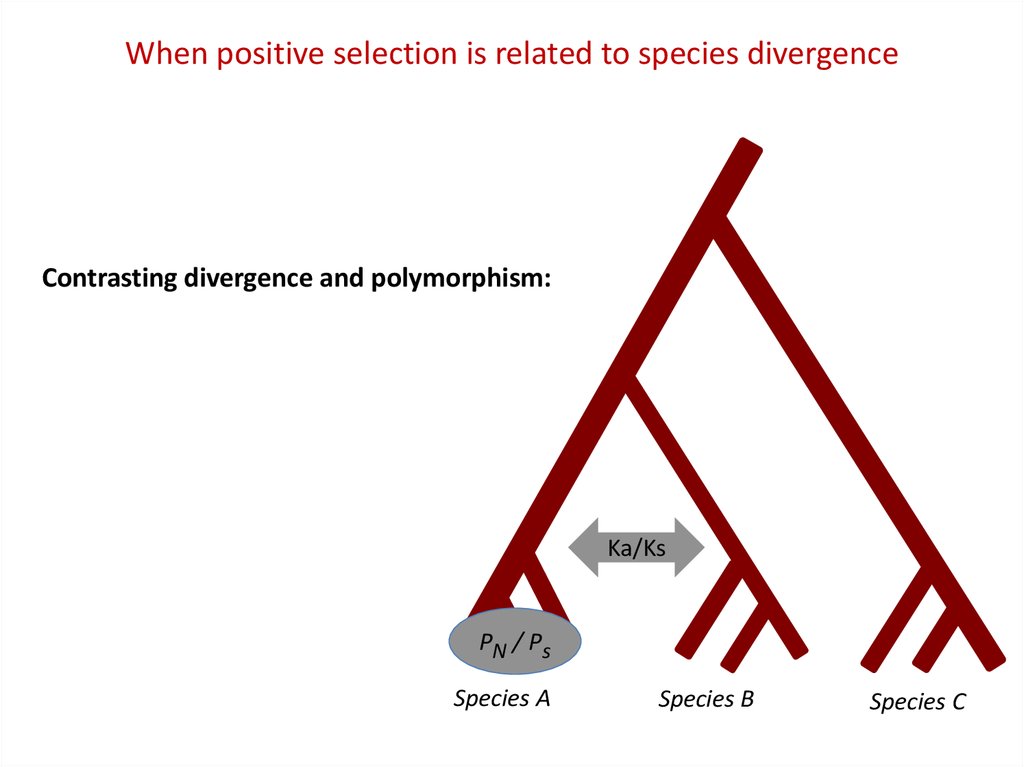
PN / Ps
Species A
Species B
Species C
15.
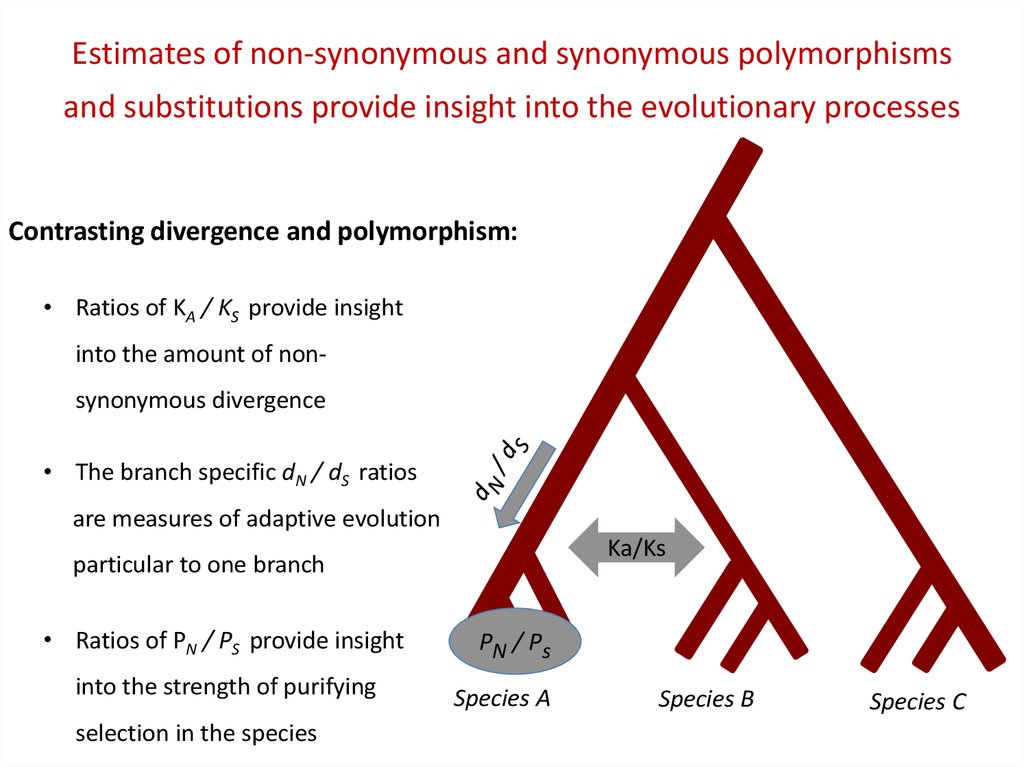
Estimates of non-synonymous and synonymous polymorphismsand substitutions provide insight into the evolutionary processes
Contrasting divergence and polymorphism:
• Ratios of KA / KS provide insight
into the amount of nonsynonymous divergence
• The branch specific dN / dS ratios
are measures of adaptive evolution
Ka/Ks
particular to one branch
• Ratios of PN / PS provide insight
into the strength of purifying
selection in the species
PN / Ps
Species A
Species B
Species C
16.
Basic analyses of the proportion of nonsynonymous to synonymous divergence KA/KS- Counts of non-synonymous mutations for each gene (Nd)
- Counts of synonymous mutations for each gene (Sd)
- Counts of potential non-synonymous sites for each gene (N)
- Counts of potential synonymous sites for each gene (S)
Non-synonymous substitution rate: KA
Synonymous substitution rate: KS
= Nd / N
= Sd / S
Ratio KA/KS as an inidicator of evolutionary
mode in each gene
Nei and Gojobori, 1986
17.
Counts of possible synonymous sites for each gene (S)Seq 1
Seq 2
1
Pro
CCC
CCC
Pro
2
Phe
UUU
UUC
Phe
Calculate potential synonymous sites (S) for each codon
A fourfold degenerate site counts as S = 1 (N = 0)
A non-degenerate site counts as S = 0 (N = 1)
A two fold degenerate site counts as S = 1/3 (N = 2/3)
1.
2.
3.
4.
5
Proline S = 0 + 0 + 1 = 1
Phenylalanine S = 0 + 0 + 1/3 = 1/3
For Glycine S = 0 + 0 + 1 = 1, for Alanine S = 0 + 0 + 1 =
Take the average: S=1
Leucine for UUA, S = 1/3 + 0 + 1/3 = 2/3
for CUA, S = 1/3 + 0 + 1 = 4/3
Take the average of these: S = 1 for codon 4
Phenylalanine for UUU, S = 1/3
for guanine, S = 1
Take average: S = 2/3
For whole sequence, S = 1 + 1/3 + 1 + 1 + 2/3 = 4
N = total number of sites: S = 15 - 4 = 11
3
Gly
GGG
GAG
Ala
4
Leu
UUA
CUA
Leu
5
Phe
UUU
GUA
Val
18.
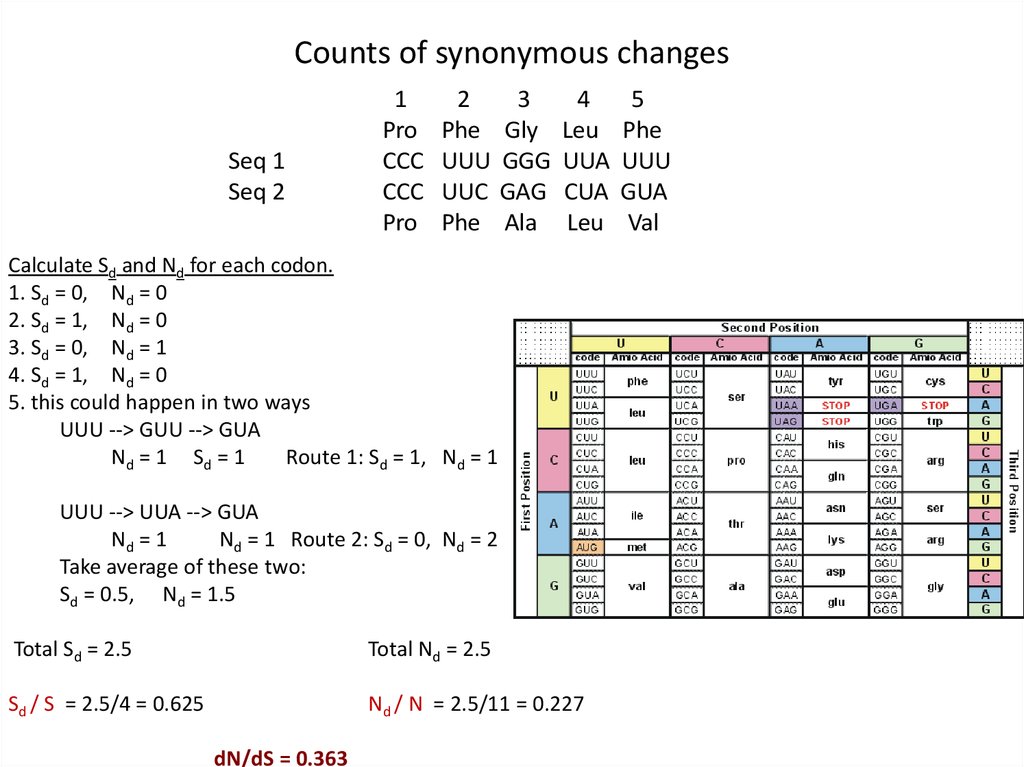
Counts of synonymous changesSeq 1
Seq 2
1
Pro
CCC
CCC
Pro
2
Phe
UUU
UUC
Phe
3
Gly
GGG
GAG
Ala
4
Leu
UUA
CUA
Leu
Calculate Sd and Nd for each codon.
1. Sd = 0, Nd = 0
2. Sd = 1, Nd = 0
3. Sd = 0, Nd = 1
4. Sd = 1, Nd = 0
5. this could happen in two ways
UUU --> GUU --> GUA
Nd = 1 Sd = 1
Route 1: Sd = 1, Nd = 1
UUU --> UUA --> GUA
Nd = 1
Nd = 1 Route 2: Sd = 0, Nd = 2
Take average of these two:
Sd = 0.5, Nd = 1.5
Total Sd = 2.5
Total Nd = 2.5
Sd / S = 2.5/4 = 0.625
Nd / N = 2.5/11 = 0.227
dN/dS = 0.363
5
Phe
UUU
GUA
Val
19.
Positive selection between speciesdN
Positive selection
Neutral evolution
f.ex. effector genes
f.ex. pseudogenes
Purifying selection
f.ex. housekeeping genes
ds
20.
When positive selection is related to species divergenceContrasting divergence and polymorphism:
Ka/Ks
PN / Ps
Species A
Species B
Species C
21.
McDonald Kreitman (MK) test to contrastwithin and between species variation
22.
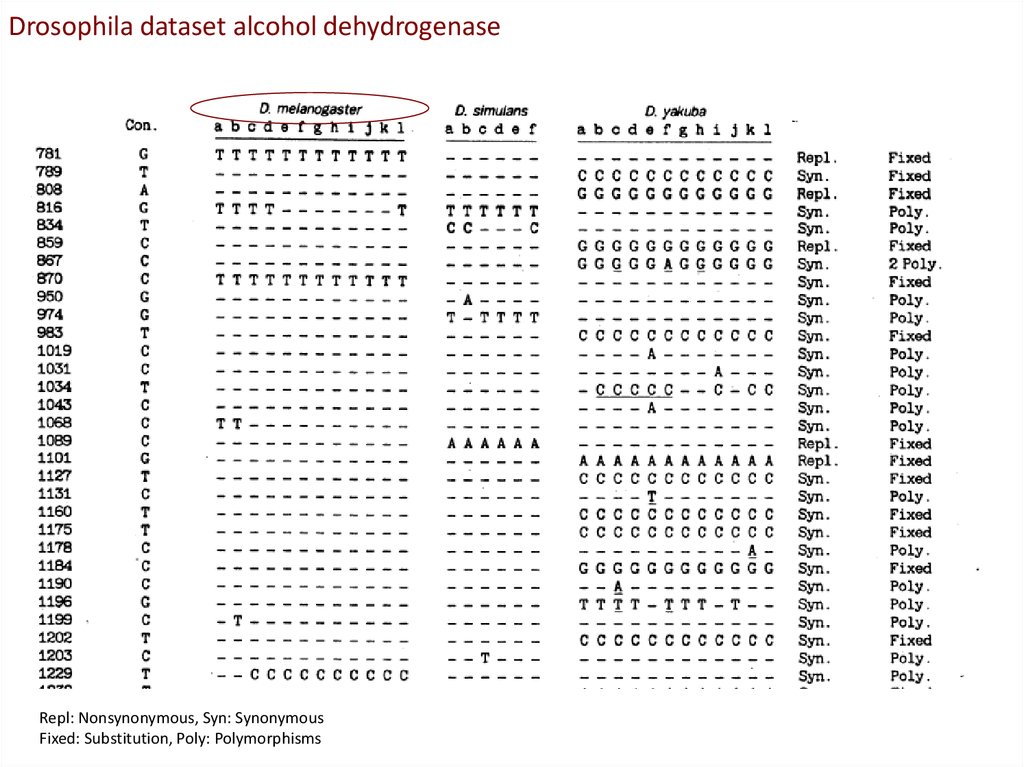
Drosophila dataset alcohol dehydrogenaseRepl: Nonsynonymous, Syn: Synonymous
Fixed: Substitution, Poly: Polymorphisms
23.
MK test contrasts within and between species synonymousand non-synonymous differences
The proportion of non-synonymous
fixed differences between species
much higher than the proportion of
non-synonymous polymorphisms
Contingency table can be tested by a G-test
24.
Conclusion from MK-test:Adh locus in Drosophila has accumulated
adaptive mutations (been under positive
selection) when the Drosophila species
diverged
25.
One problem with the “counting methods”Sometimes the signal of selection is not very strong
26.
Positive selection on one or few particular codonsor in one particular branch
Evolutionary model to detect selection in particular codons or branches














 Биология
Биология








