Похожие презентации:
Microbial Metabolism. Fueling Cell Growth
1.
Microbial MetabolismFueling Cell Growth
2.
A Glimpse of HistoryBiologists had noticed that in vats of grape juice,
alcohol and CO2 are produced while yeast cells
increase in number
• But idea not widely accepted, mocked by some
• In 1850s, Louis Pasteur set out to prove
• Simplified setup: clear solution of sugar, ammonia,
mineral salts, trace elements
• Added a few yeast cells—as they grew, sugar decreased,
alcohol level increased
• Strongly supported idea, but Pasteur failed to extract
something from inside the cells that would convert sugar
• In 1897, Eduard Buchner, a German chemist, showed
that crushed yeast cells could convert sugar to ethanol
and CO2; awarded Nobel Prize in 1907
3.
Learning OutcomesCompare and contrast catabolism and anabolism.
Describe the energy sources used by photosynthetic
organisms
and chemoorganoheterotrophs.
Describe the components of metabolic pathways
(enzymes, ATP, chemical energy sources and terminal
electron acceptors, and electron carriers) and the role
of precursor metabolites.
Describe the roles of the three central metabolic
pathways.
Distinguish between respiration and fermentation.
4.
Microbial MetabolismAll cells need to accomplish two fundamental tasks
• Synthesize new parts
• Cell walls, membranes, ribosomes, nucleic acids
• Harvest energy to power reactions
Sum total of chemical reaction in a cell is called metabolism
Implications of microbial metabolism
• Biofuels
• Food production
• Important in laboratory
• Invaluable models for study
• Unique pathways potential
drug targets
5.
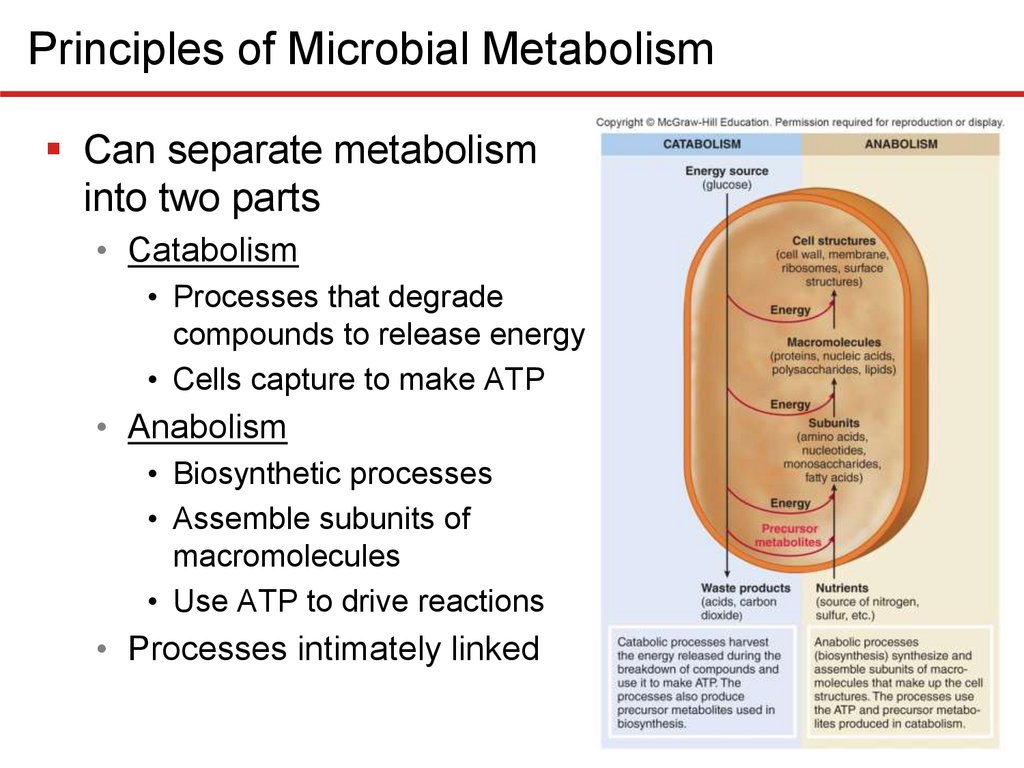
Principles of Microbial MetabolismCan separate metabolism
into two parts
• Catabolism
• Processes that degrade
compounds to release energy
• Cells capture to make ATP
• Anabolism
• Biosynthetic processes
• Assemble subunits of
macromolecules
• Use ATP to drive reactions
• Processes intimately linked
6.
EnergyEnergy is the capacity to do work
Two types of energy
• Potential: stored energy (e.g., chemical bonds, rock on
hill, water behind dam)
• Kinetic: energy of movement (e.g., moving water)
• Energy in universe cannot be
created or destroyed, but it can
be converted between forms
7.
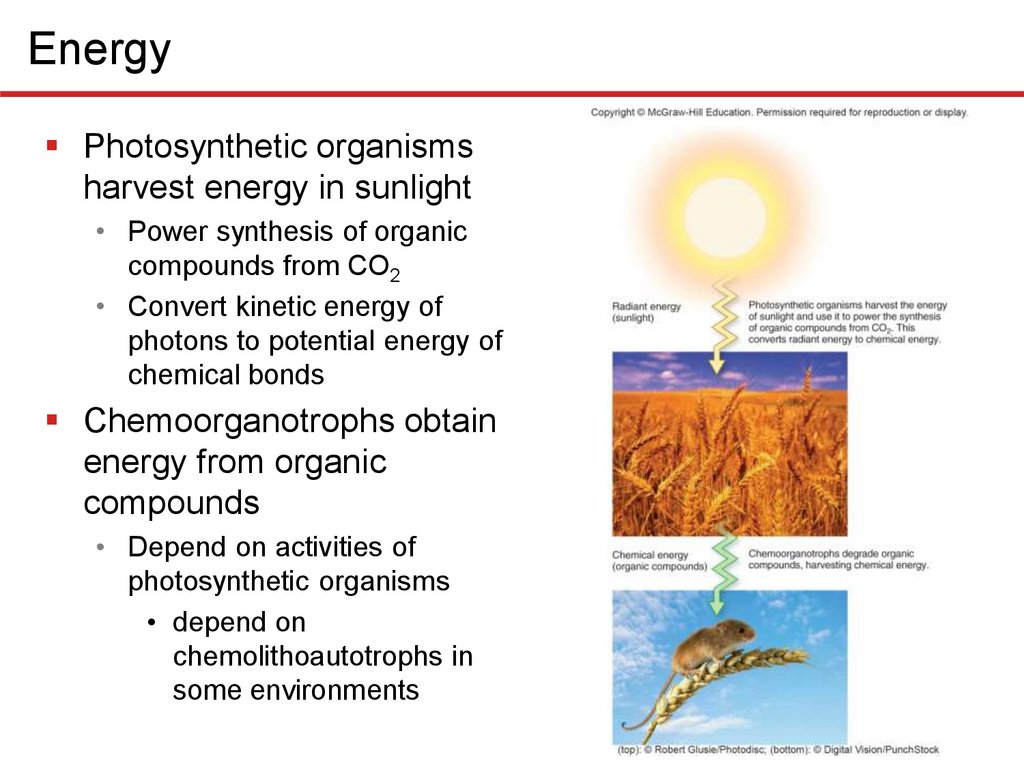
EnergyPhotosynthetic organisms
harvest energy in sunlight
• Power synthesis of organic
compounds from CO2
• Convert kinetic energy of
photons to potential energy of
chemical bonds
Chemoorganotrophs obtain
energy from organic
compounds
• Depend on activities of
photosynthetic organisms
• depend on
chemolithoautotrophs in
some environments
8.
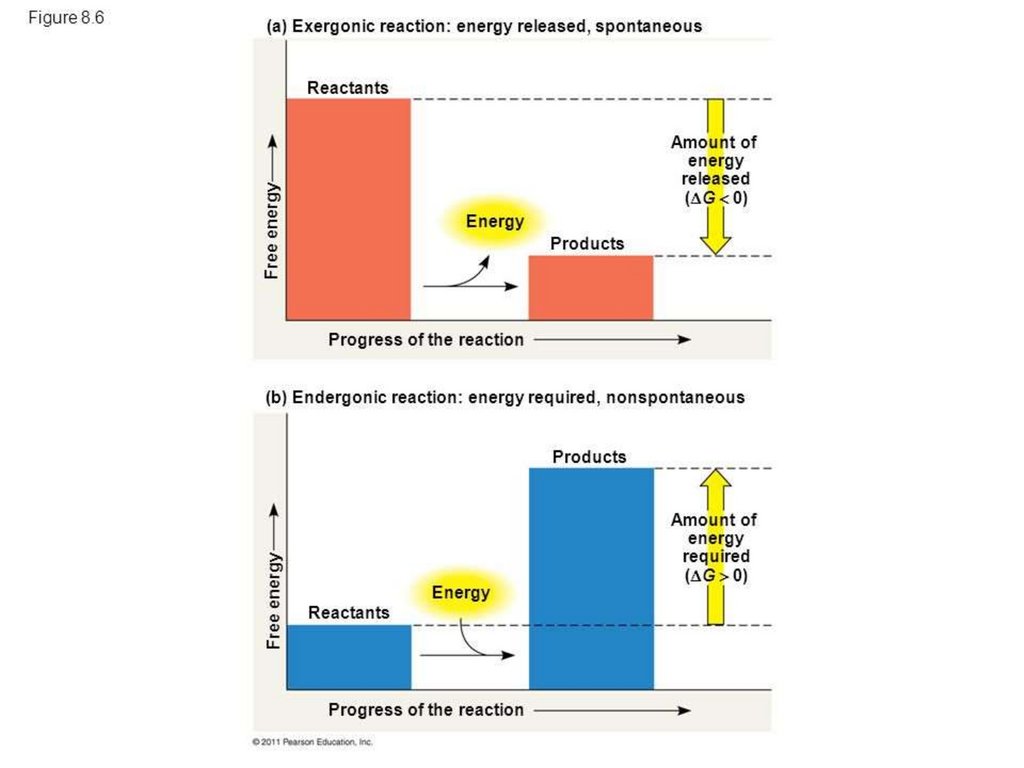
EnergyFree energy is energy available to do work
• E.g., energy released when chemical bond is broken
• Compare free energy of reactants, products
• Exergonic reactions: reactants have more free energy
• Energy is released in reaction
• Endergonic reactions: products have more free energy
• Reaction requires input of energy
• Change in free energy is same regardless of number of
steps involved (e.g., converting glucose to CO2 + H2O)
• Cells use multiple steps when degrading compounds
• Energy released from exergonic reactions powers
endergonic reactions
9.
10.
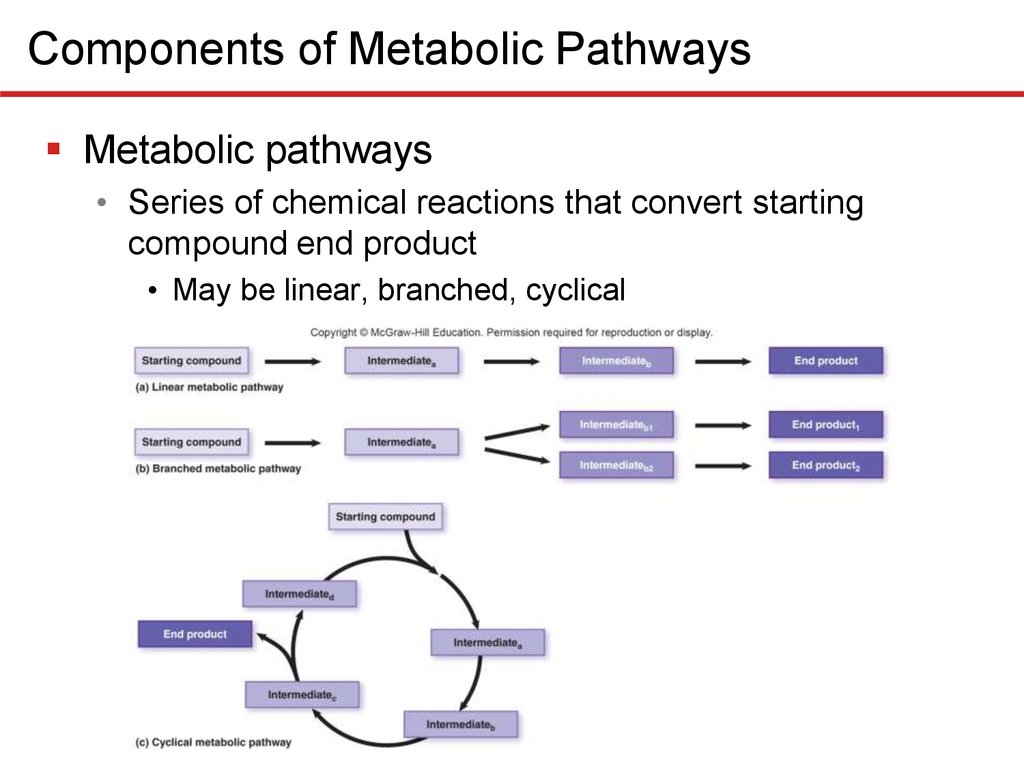
Components of Metabolic PathwaysMetabolic pathways
• Series of chemical reactions that convert starting
compound end product
• May be linear, branched, cyclical
11.
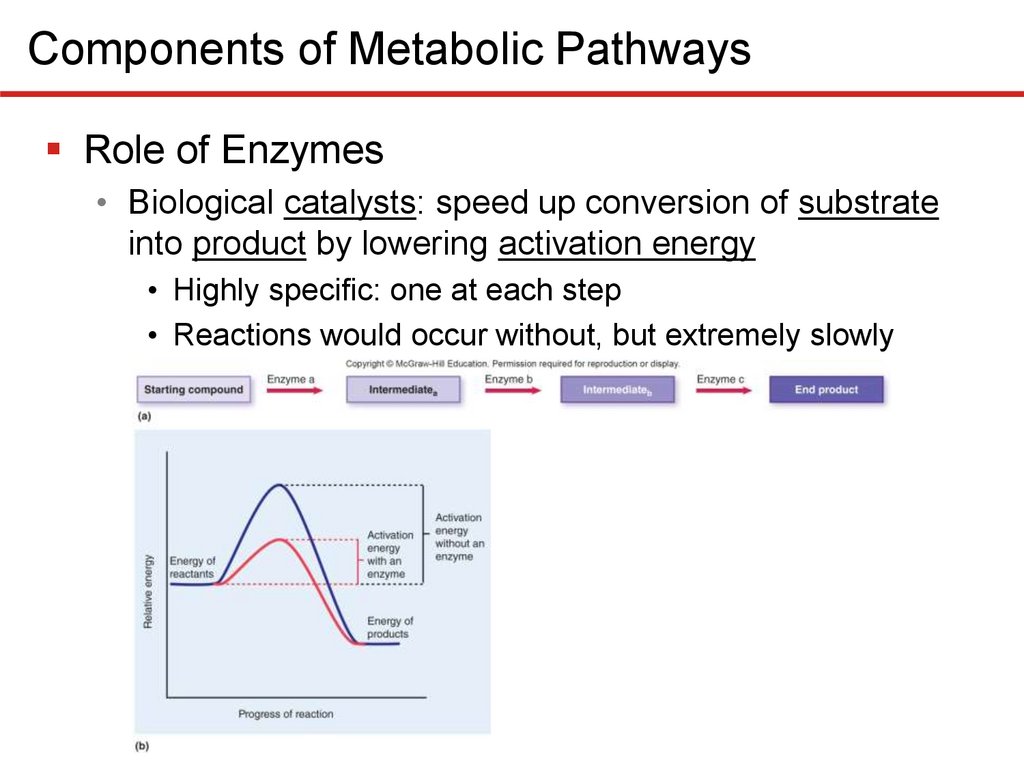
Components of Metabolic PathwaysRole of Enzymes
• Biological catalysts: speed up conversion of substrate
into product by lowering activation energy
• Highly specific: one at each step
• Reactions would occur without, but extremely slowly
12.
Q&AWhat would happen if there were no enzymes in the
body?
13.
Components of Metabolic PathwaysRole of ATP
• Adenosine triphospate (ATP) is energy currency of the
cell
• Composed of ribose, adenine, three phosphate groups
• Adenosine diphospate (ADP) acceptor of free energy
• Cells produce ATP by adding Pi to ADP using energy
• Release energy from ATP to yield ADP and Pi
Three processes to generate ATP
• Substrate-level phosphorylation
• Exergonic reaction
• Oxidative phosphorylation
• Proton motive force
• Photophosphorylation
• Sunlight used to create proton
motive force
14.
Components of Metabolic PathwaysRole of the Chemical Energy Source and the
Terminal Electron Acceptor
Some atoms, molecules more electronegative
than others
• Greater affinity for electrons
• Energy released when
electrons move from low
affinity molecule to high
affinity molecule
• (glucose to O2)
15.
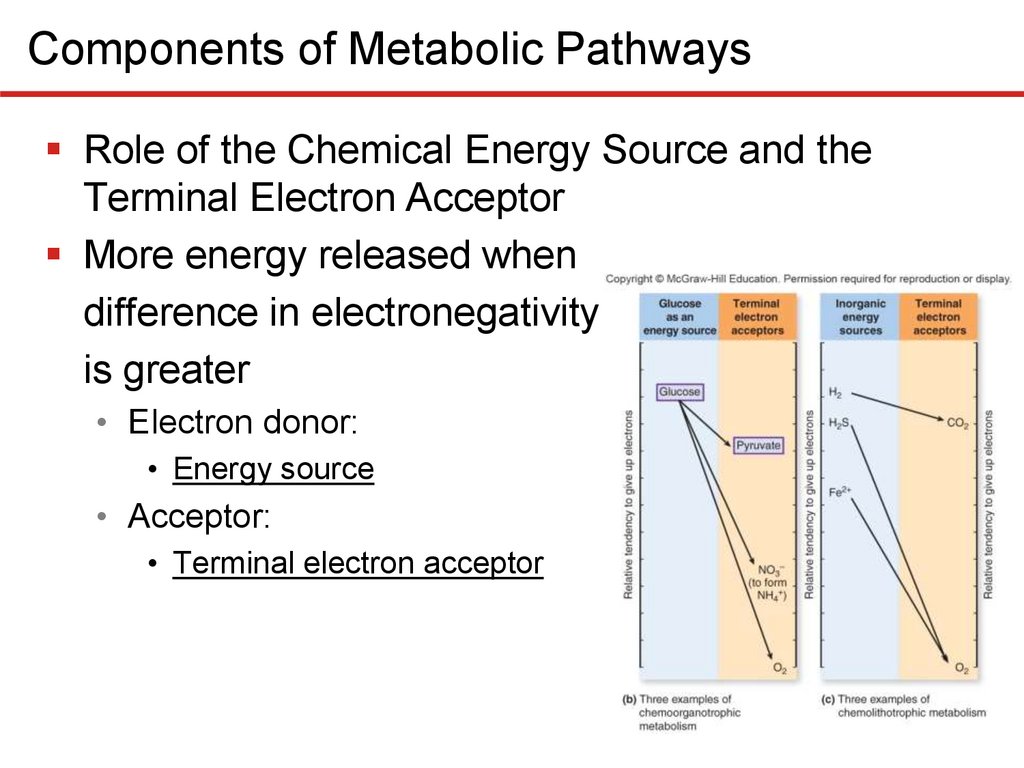
Components of Metabolic PathwaysRole of the Chemical Energy Source and the
Terminal Electron Acceptor
More energy released when
difference in electronegativity
is greater
• Electron donor:
• Energy source
• Acceptor:
• Terminal electron acceptor
16.
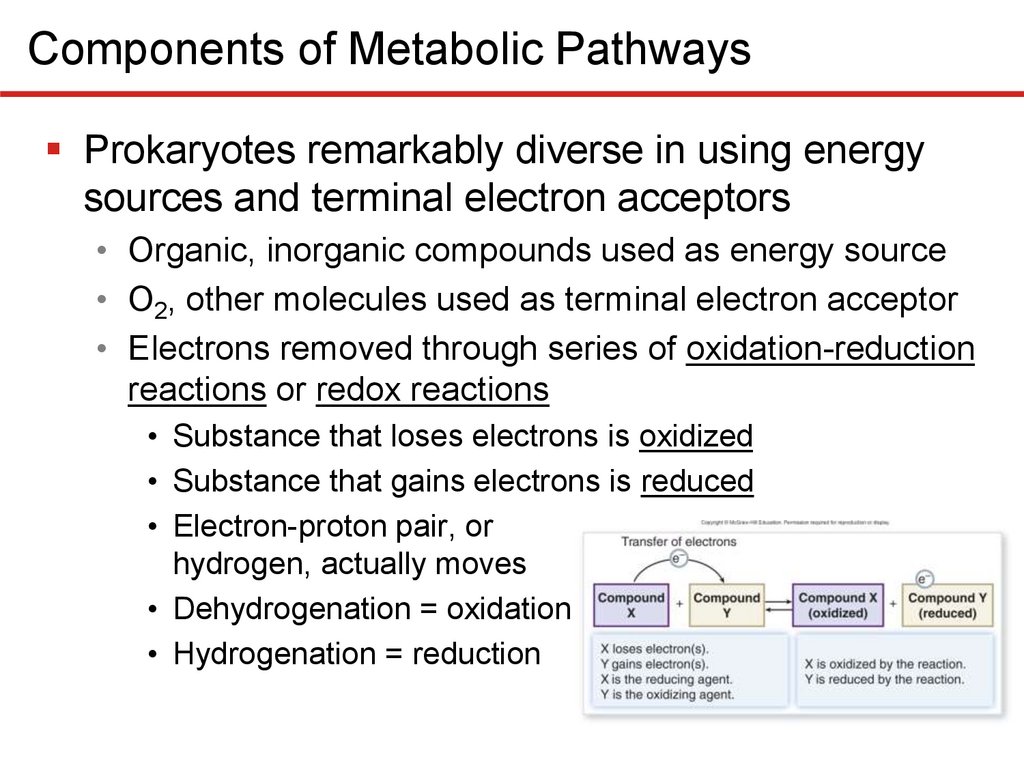
Components of Metabolic PathwaysProkaryotes remarkably diverse in using energy
sources and terminal electron acceptors
• Organic, inorganic compounds used as energy source
• O2, other molecules used as terminal electron acceptor
• Electrons removed through series of oxidation-reduction
reactions or redox reactions
• Substance that loses electrons is oxidized
• Substance that gains electrons is reduced
• Electron-proton pair, or
hydrogen, actually moves
• Dehydrogenation = oxidation
• Hydrogenation = reduction
17.
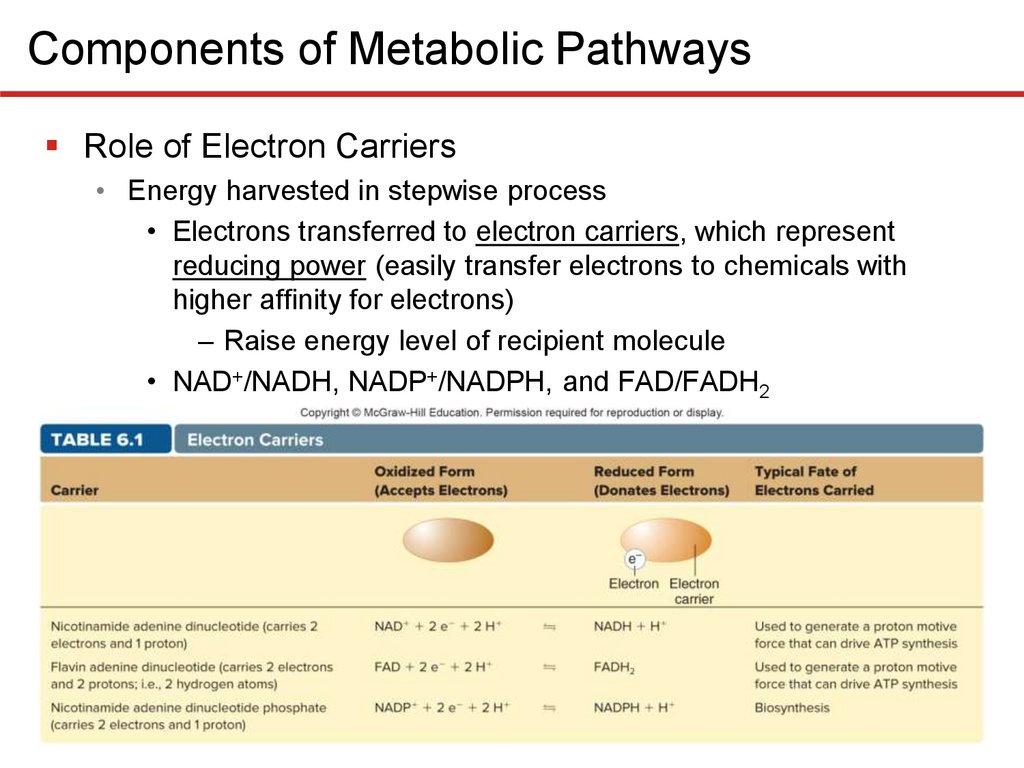
Components of Metabolic PathwaysRole of Electron Carriers
• Energy harvested in stepwise process
• Electrons transferred to electron carriers, which represent
reducing power (easily transfer electrons to chemicals with
higher affinity for electrons)
– Raise energy level of recipient molecule
• NAD+/NADH, NADP+/NADPH, and FAD/FADH2
18.
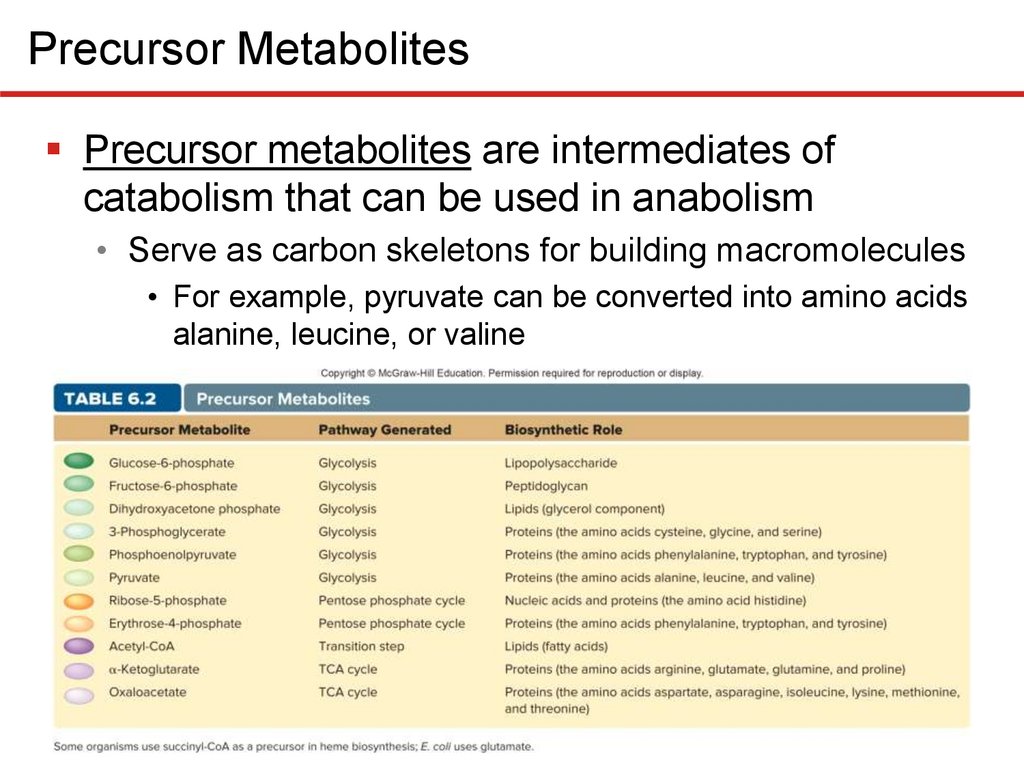
Precursor MetabolitesPrecursor metabolites are intermediates of
catabolism that can be used in anabolism
• Serve as carbon skeletons for building macromolecules
• For example, pyruvate can be converted into amino acids
alanine, leucine, or valine
19.
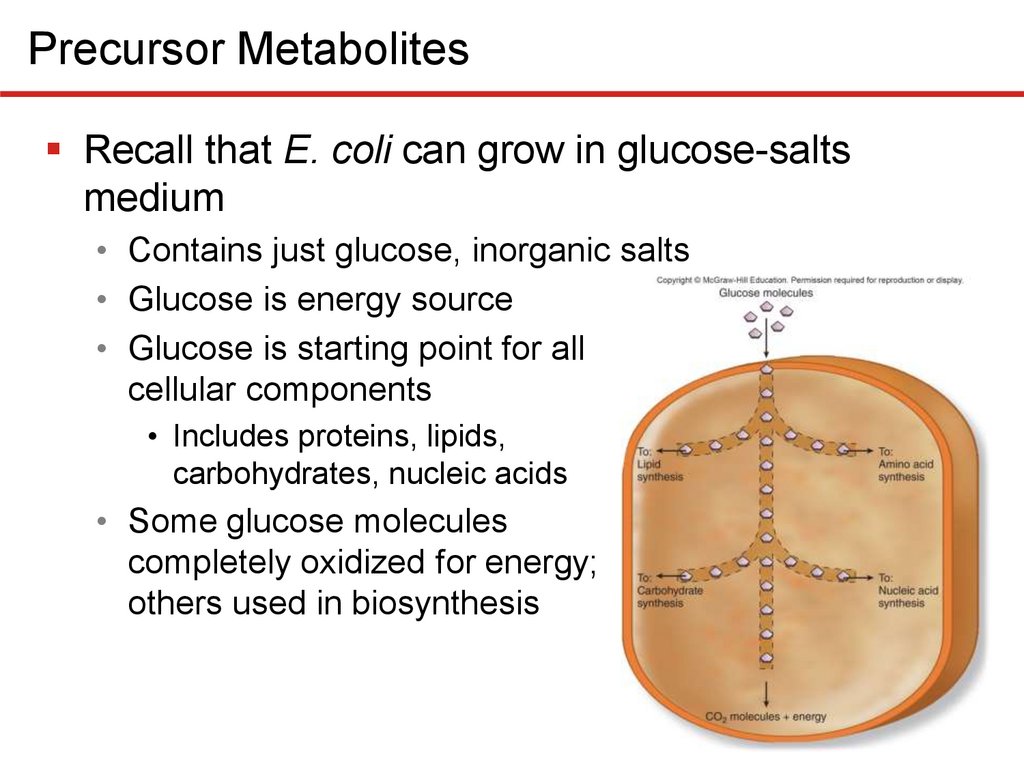
Precursor MetabolitesRecall that E. coli can grow in glucose-salts
medium
• Contains just glucose, inorganic salts
• Glucose is energy source
• Glucose is starting point for all
cellular components
• Includes proteins, lipids,
carbohydrates, nucleic acids
• Some glucose molecules
completely oxidized for energy;
others used in biosynthesis
20.
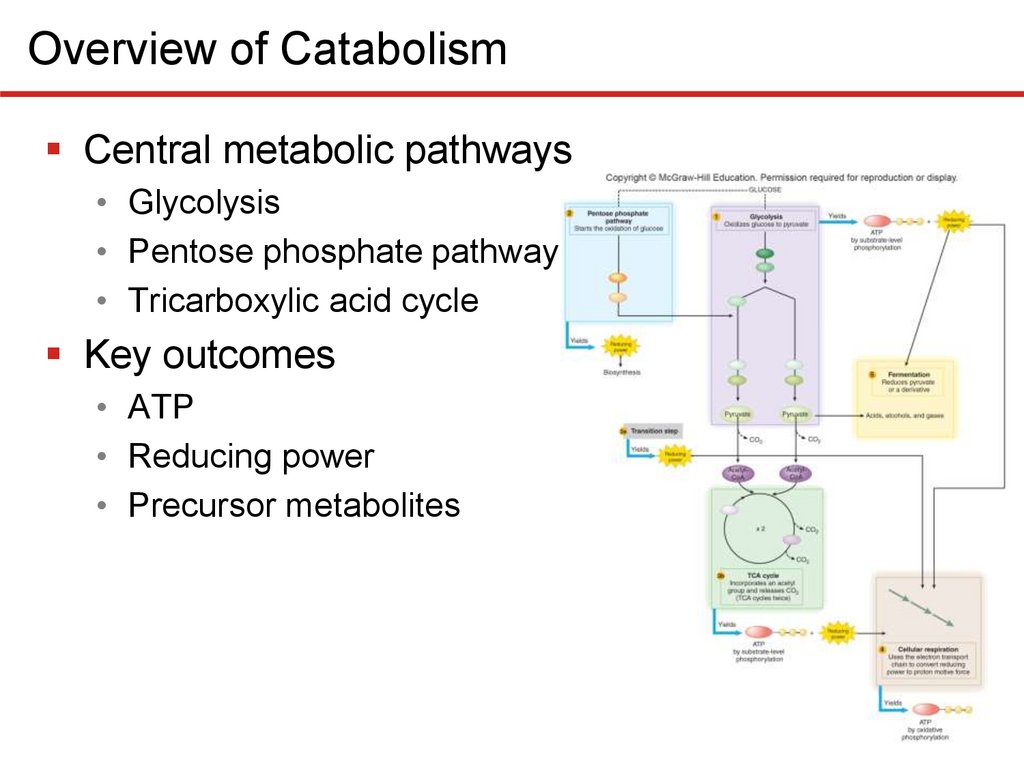
Overview of CatabolismThree central metabolic pathways
• Oxidize glucose to CO2
• Catabolic and precursor metabolites as well a reducing
power can be diverted for use in biosynthesis
• Termed amphibolic to reflect dual role
• Glycolysis
• Splits glucose (6C) to two pyruvates (3C)
• Generates modest ATP, reducing power, precursors
• Pentose phosphate pathway
• Primary role is production precursor metabolites, NADPH
• Tricarboxylic acid cycle
• Oxidizes pyruvates from glycolysis
• Generates reducing power, precursor metabolites, ATP
21.
Overview of CatabolismCentral metabolic pathways
• Glycolysis
• Pentose phosphate pathway
• Tricarboxylic acid cycle
Key outcomes
• ATP
• Reducing power
• Precursor metabolites
22.
Overview of CatabolismRespiration transfers electrons from glucose to
electron transport chain
• Electron transport chain generates proton motive force
• Harvested to make ATP via oxidative phosphorylation
• Aerobic respiration
– O2 is terminal electron acceptor
• Anaerobic respiration
– Molecule other than O2 as terminal electron acceptor
– Also use modified version of TCA cycle
23.
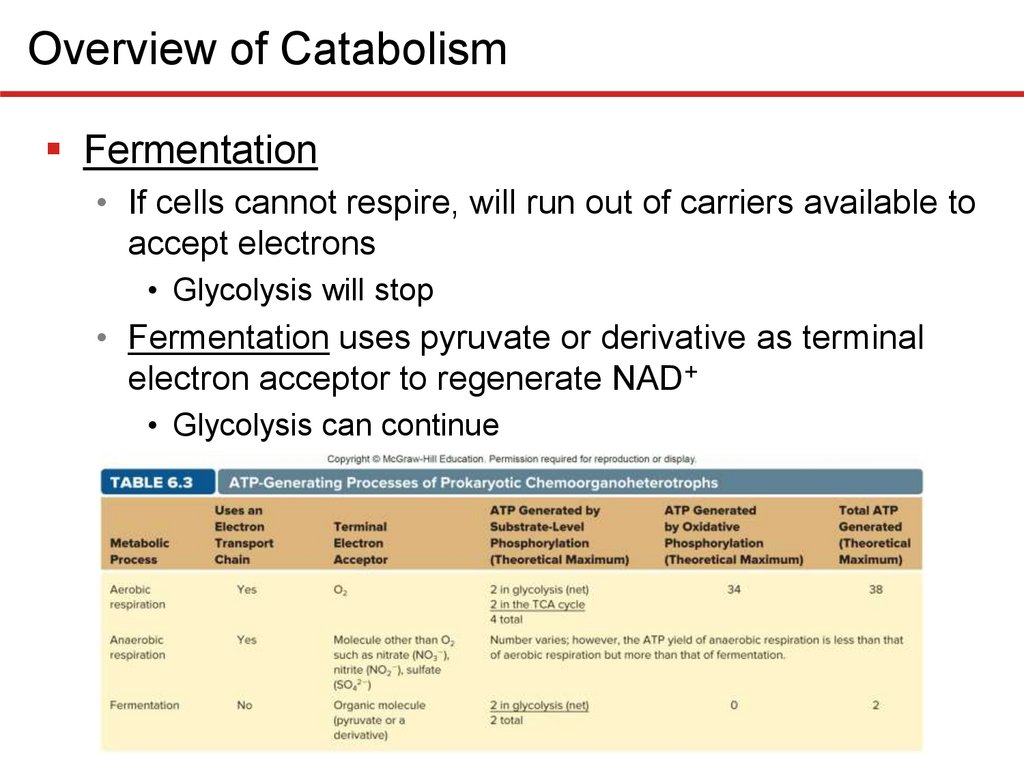
Overview of CatabolismFermentation
• If cells cannot respire, will run out of carriers available to
accept electrons
• Glycolysis will stop
• Fermentation uses pyruvate or derivative as terminal
electron acceptor to regenerate NAD+
• Glycolysis can continue
24.
2 EnzymesDescribe the active site of an enzyme, and
explain how it relates to the enzyme-substrate
complex.
Compare and contrast cofactors and
coenzymes.
List two environmental factors that influence
enzyme activity.
Describe allosteric regulation.
Compare and contrast competitive enzyme
inhibition and non-competitive enzyme
inhibition.
25.
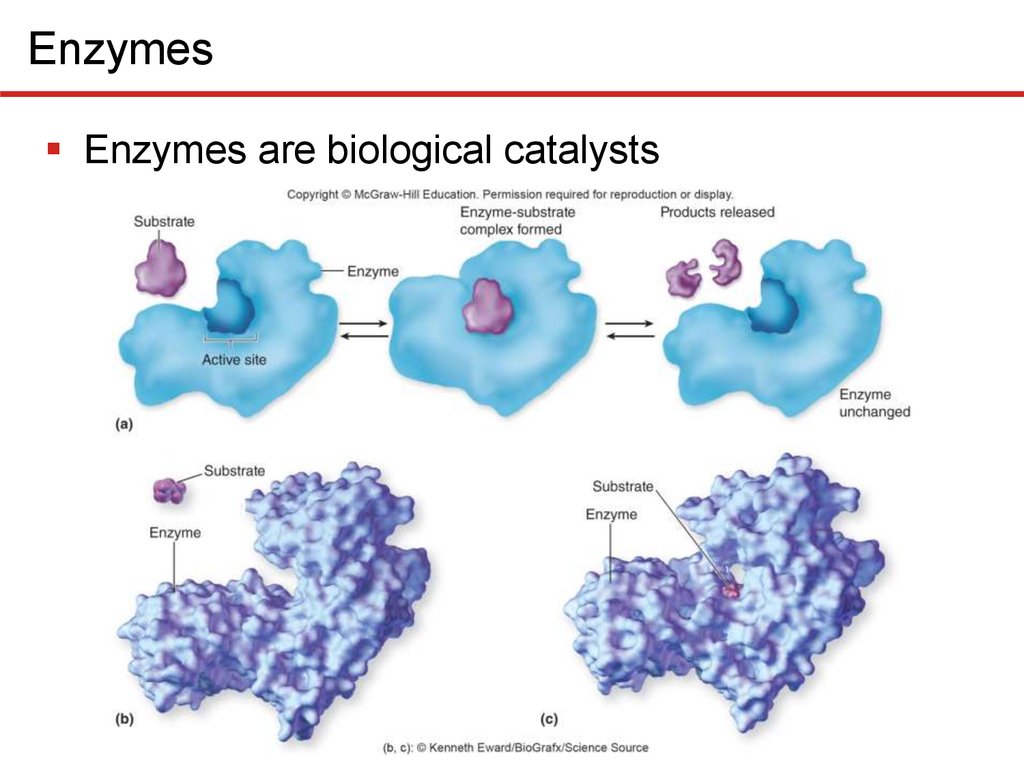
2 EnzymesEnzymes are biological catalysts
• Name reflects function; ends in -ase
• Has active site to which substrate(s) bind(s) weakly
• Causes enzyme shape to change slightly, induced fit
• Existing substrate bonds destabilized, new ones form
• Enzymes are highly specific for substrate(s)
• Enzyme not used up
26.
EnzymesEnzymes are biological catalysts
27.
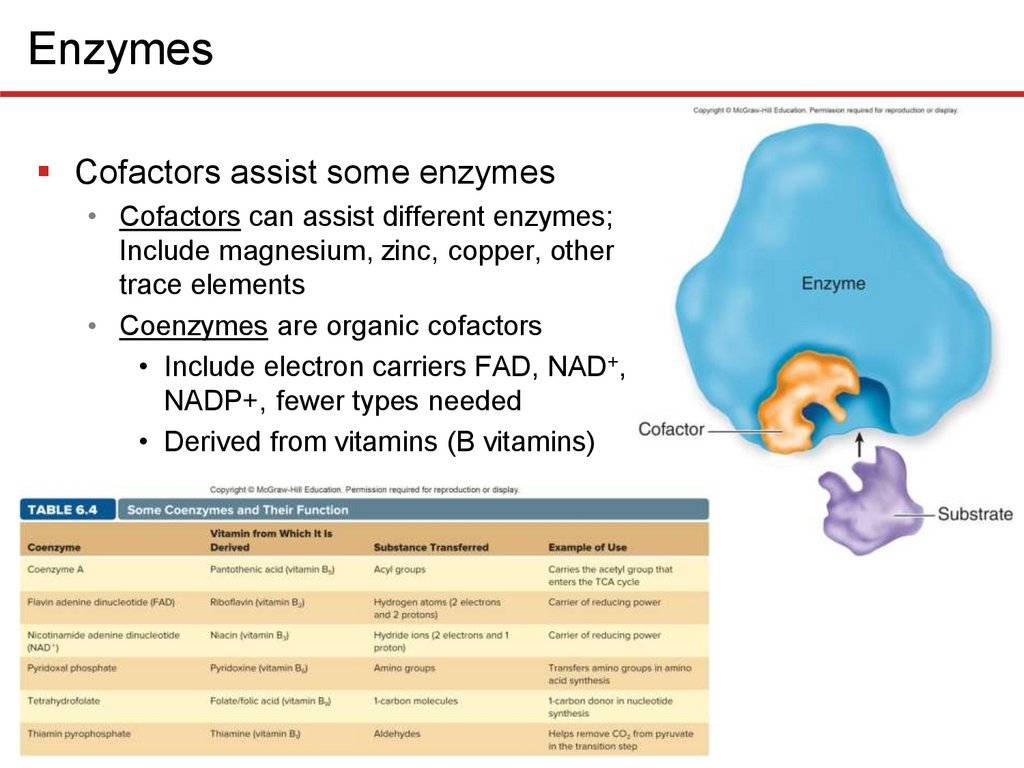
EnzymesCofactors assist some enzymes
• Cofactors can assist different enzymes;
Include magnesium, zinc, copper, other
trace elements
• Coenzymes are organic cofactors
• Include electron carriers FAD, NAD+,
NADP+, fewer types needed
• Derived from vitamins (B vitamins)
28.
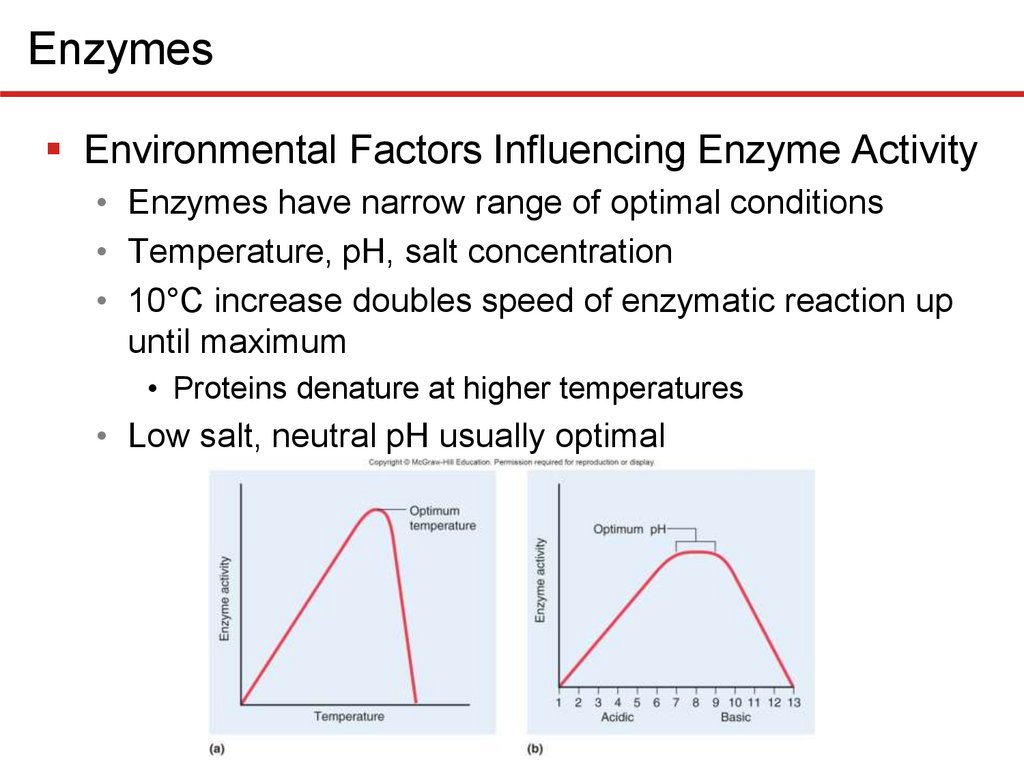
EnzymesEnvironmental Factors Influencing Enzyme Activity
• Enzymes have narrow range of optimal conditions
• Temperature, pH, salt concentration
• 10°C increase doubles speed of enzymatic reaction up
until maximum
• Proteins denature at higher temperatures
• Low salt, neutral pH usually optimal
29.
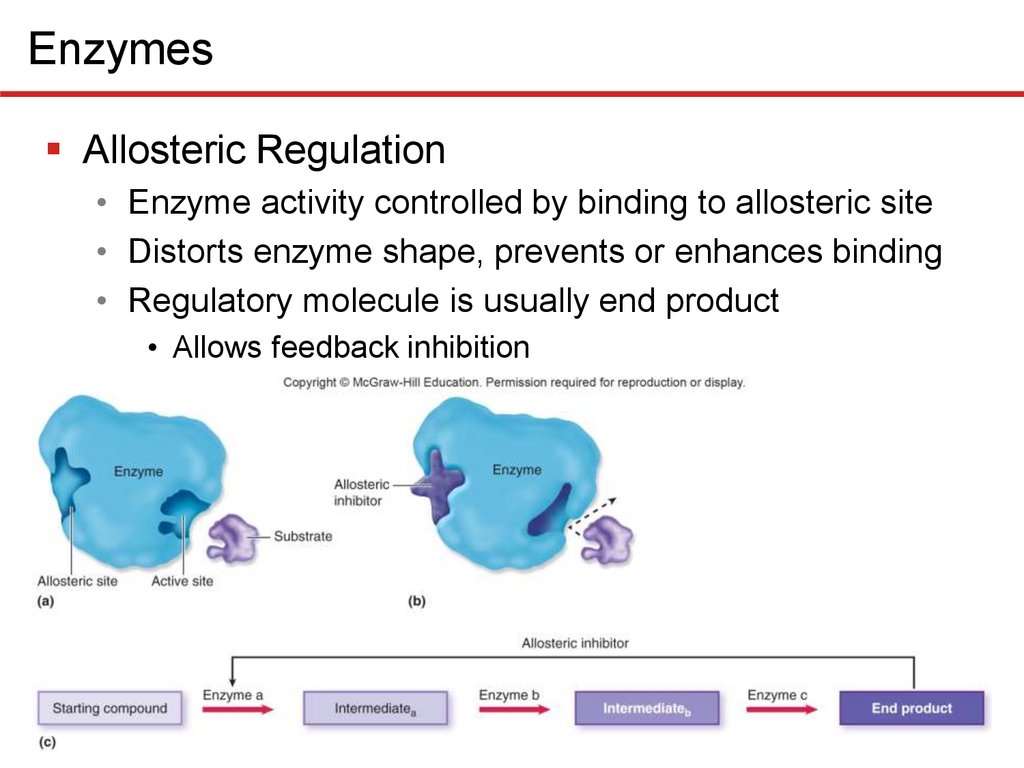
EnzymesAllosteric Regulation
• Enzyme activity controlled by binding to allosteric site
• Distorts enzyme shape, prevents or enhances binding
• Regulatory molecule is usually end product
• Allows feedback inhibition
30.
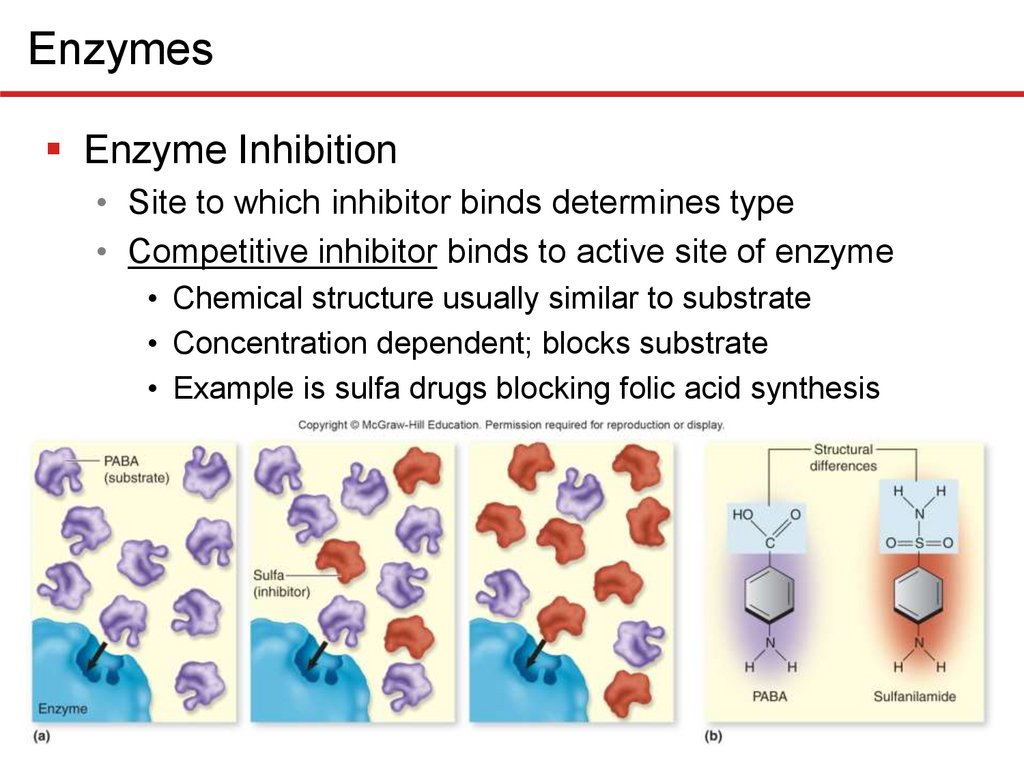
EnzymesEnzyme Inhibition
• Site to which inhibitor binds determines type
• Competitive inhibitor binds to active site of enzyme
• Chemical structure usually similar to substrate
• Concentration dependent; blocks substrate
• Example is sulfa drugs blocking folic acid synthesis
31.
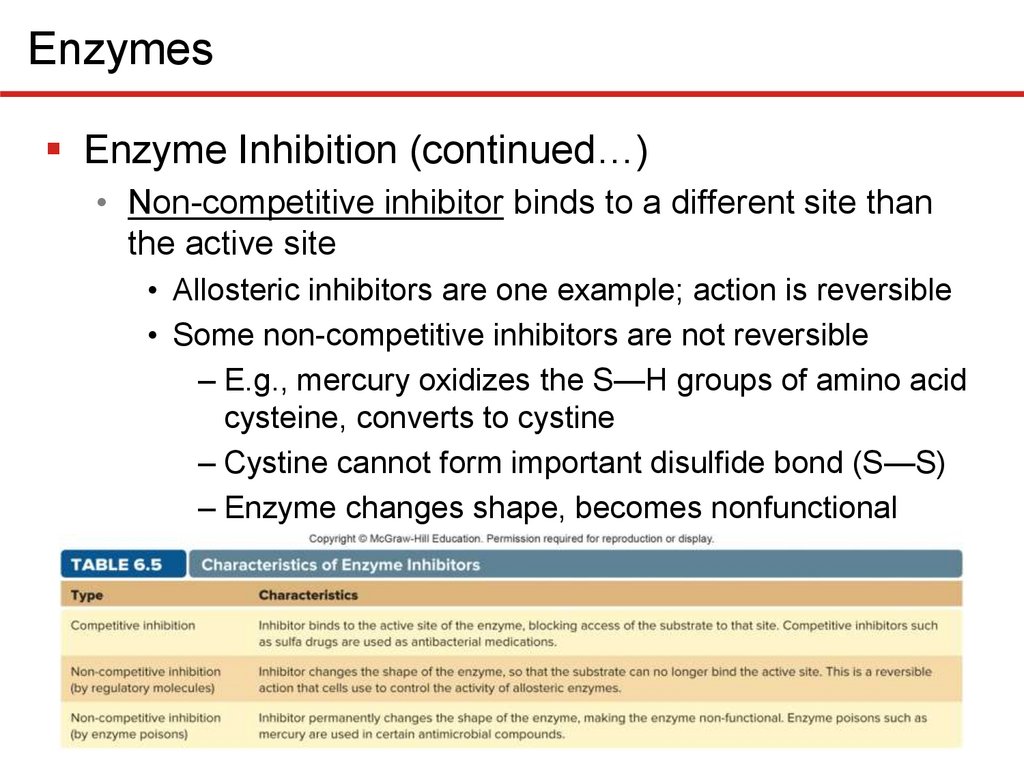
EnzymesEnzyme Inhibition (continued…)
• Non-competitive inhibitor binds to a different site than
the active site
• Allosteric inhibitors are one example; action is reversible
• Some non-competitive inhibitors are not reversible
– E.g., mercury oxidizes the S—H groups of amino acid
cysteine, converts to cystine
– Cystine cannot form important disulfide bond (S—S)
– Enzyme changes shape, becomes nonfunctional
32.
3. The Central Metabolic PathwaysATP
Reducing power: NADH, FADH2, NADPH
Precursor metabolites
• Glucose molecules can have
different fates
• Can be completely oxidized
to CO2 for maximum ATP
• Can be siphoned off as
precursor metabolite for
use in biosynthesis
33.
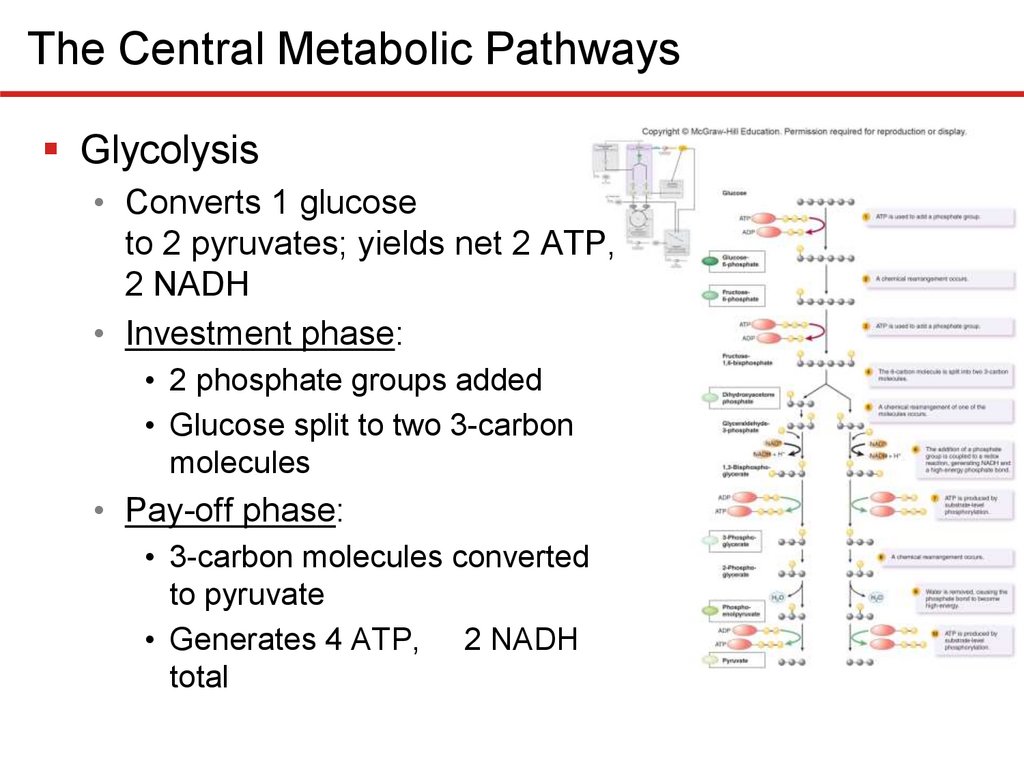
The Central Metabolic PathwaysGlycolysis
• Converts 1 glucose
to 2 pyruvates; yields net 2 ATP,
2 NADH
• Investment phase:
• 2 phosphate groups added
• Glucose split to two 3-carbon
molecules
• Pay-off phase:
• 3-carbon molecules converted
to pyruvate
• Generates 4 ATP, 2 NADH
total
34.
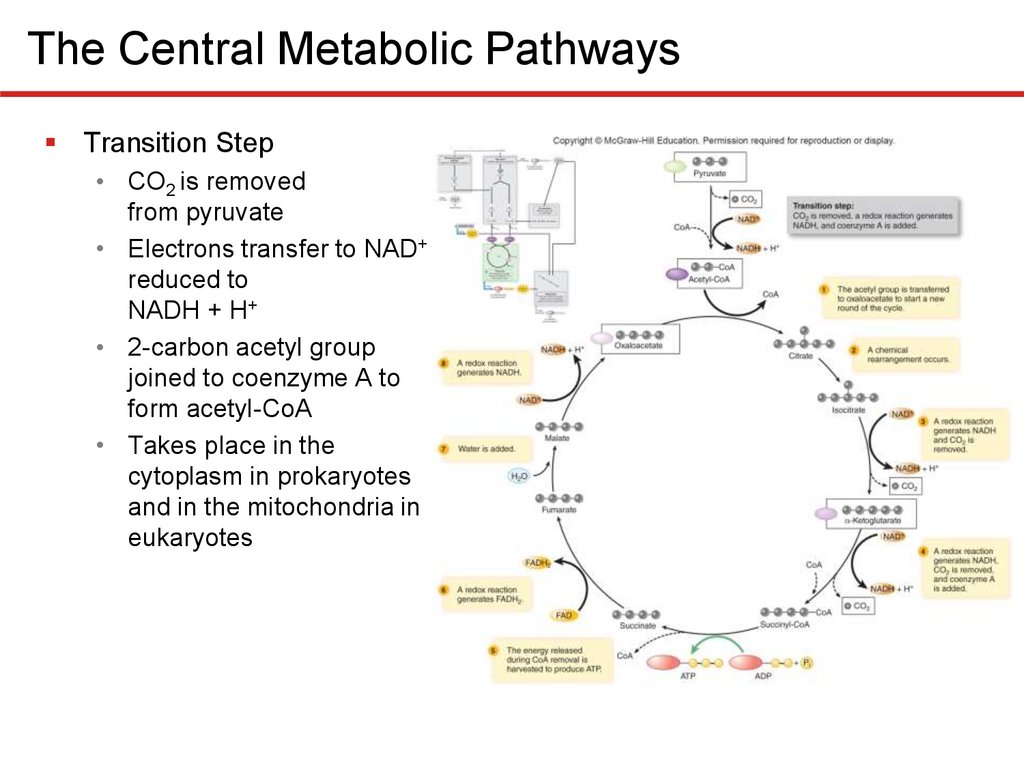
The Central Metabolic PathwaysTransition Step
• CO2 is removed
from pyruvate
• Electrons transfer to NAD+
reduced to
NADH + H+
• 2-carbon acetyl group
joined to coenzyme A to
form acetyl-CoA
• Takes place in the
cytoplasm in prokaryotes
and in the mitochondria in
eukaryotes
35.
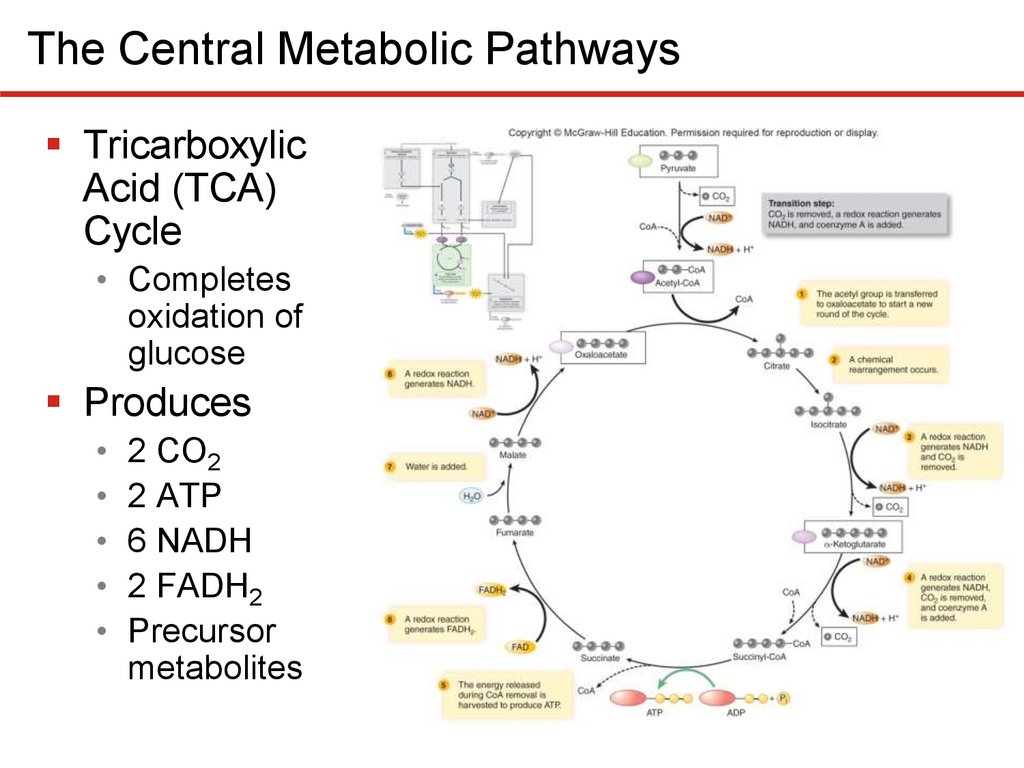
The Central Metabolic PathwaysTricarboxylic
Acid (TCA)
Cycle
• Completes
oxidation of
glucose
Produces
• 2 CO2
• 2 ATP
• 6 NADH
• 2 FADH2
• Precursor
metabolites
36.
4. RespirationUses reducing power (NADH, FADH2) generated
by glycolysis, transition step, and TCA cycle to
synthesize ATP
• Electron transport chain generates proton motive force
• Drives synthesis of ATP by ATP synthase
• Process proposed by British scientist Peter Mitchell in
1961
• Initially widely dismissed
• Mitchell conducted years of self-funded research
• Received a Nobel Prize in 1978
• Now called chemiosmotic theory
37.
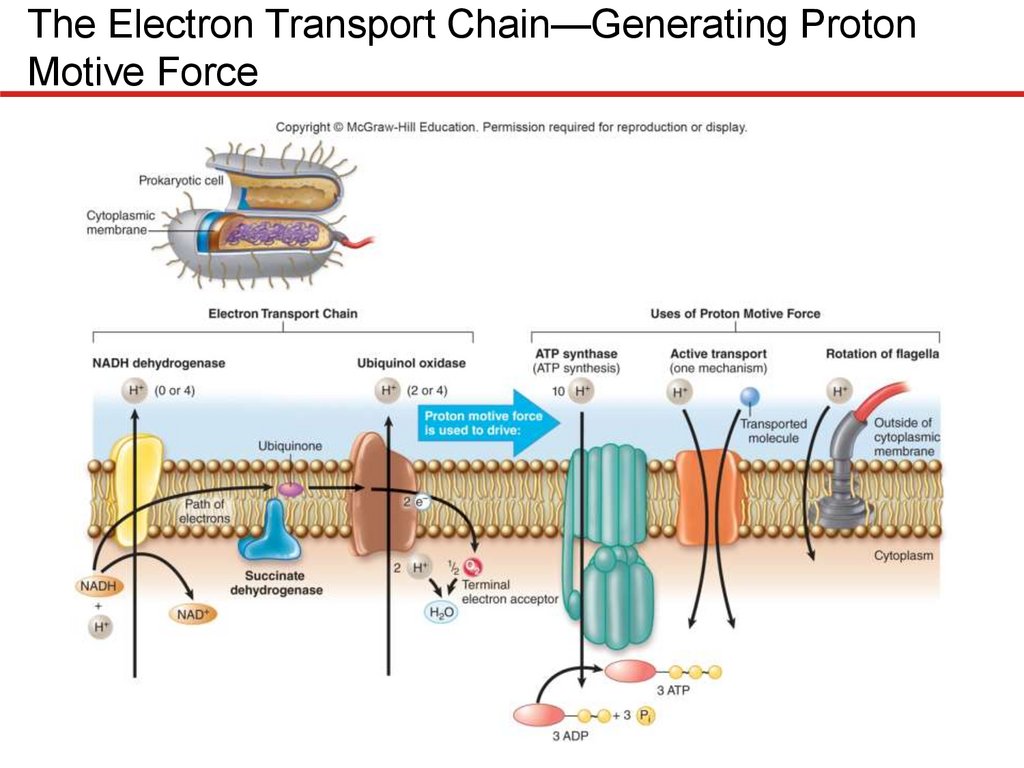
The Electron Transport Chain—Generating ProtonMotive Force
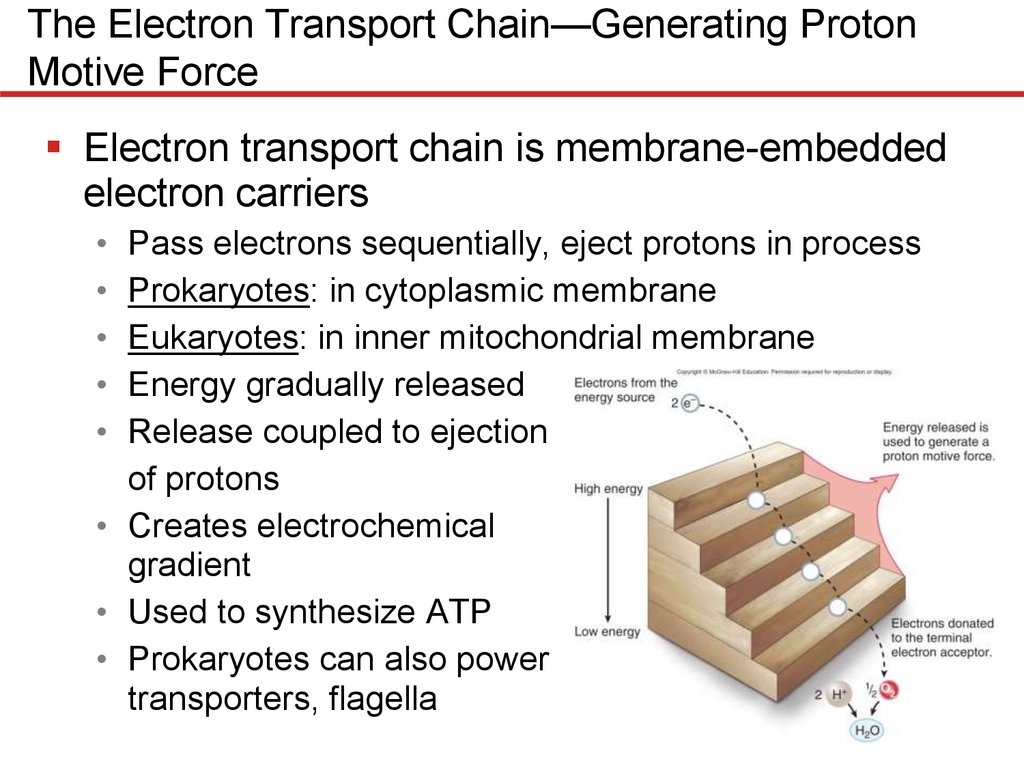
Electron transport chain is membrane-embedded
electron carriers
• Pass electrons sequentially, eject protons in process
• Prokaryotes: in cytoplasmic membrane
• Eukaryotes: in inner mitochondrial membrane
• Energy gradually released
• Release coupled to ejection
of protons
• Creates electrochemical
gradient
• Used to synthesize ATP
• Prokaryotes can also power
transporters, flagella
38.
The Electron Transport Chain—Generating ProtonMotive Force
Components of an Electron Transport Chain
• Most carriers grouped into large protein complexes
• Serve as proton pumps
• Three general groups are notable
• Quinones
• Lipid-soluble molecules
• Move freely, can transfer electrons between complexes
• Cytochromes
• Contain heme, molecule with iron atom at center
• Several types
• Flavoproteins
• Proteins to which a flavin is attached
• FAD, other flavins synthesized from riboflavin
39.
The Electron Transport Chain—Generating ProtonMotive Force
General Mechanisms of Proton Ejection
• Some carriers accept only hydrogen atoms (protonelectron pairs), others only electrons
• Spatial arrangement in membrane shuttles protons to
outside of membrane
• When hydrogen carrier accepts electron from electron
carrier, it picks up proton from inside cell
– or mitochondrial matrix
• When hydrogen carrier passes electrons to electron
carrier, protons released to outside of cell
– or intermembrane space of mitochondria
• Net effect is movement of protons across membrane
• Establishes concentration gradient
• Driven by energy released during electron transfer
40.
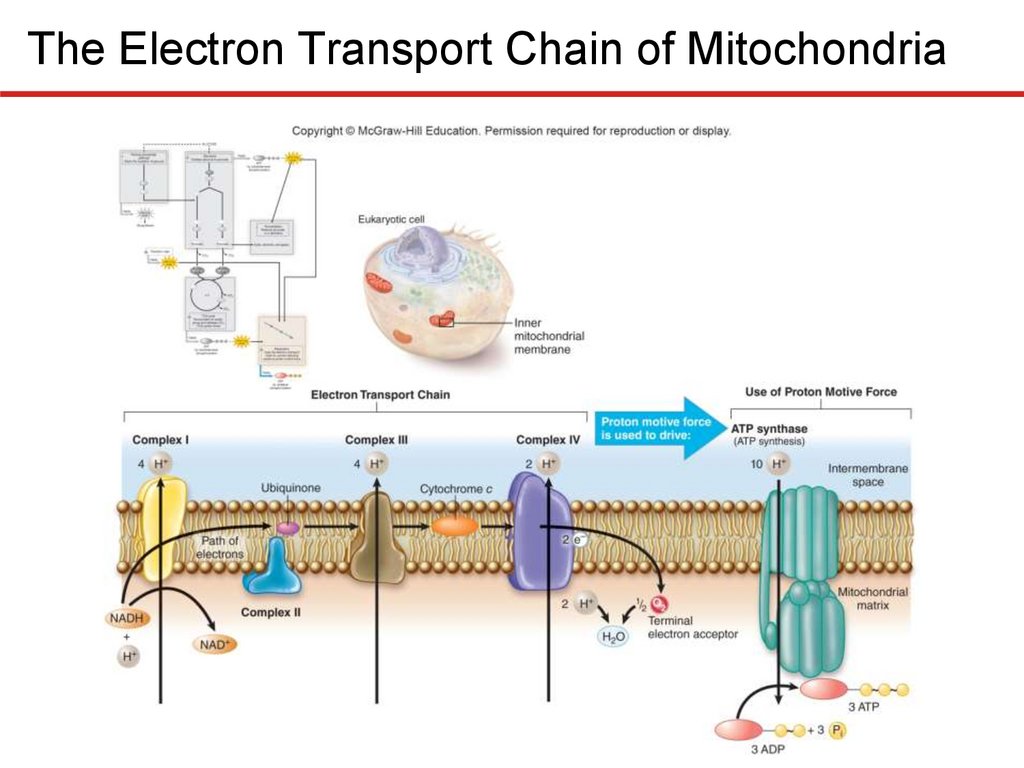
The Electron Transport Chain—Generating ProtonMotive Force
Electron Transport Chain of Mitochondria
• Complex I (NADH dehydrogenase complex)
• Accepts electrons from NADH, transfers to ubiquinone
• Pumps 4 protons
• Complex II (succinate dehydrogenase complex)
• Accepts electrons from TCA cycle via FADH2, “downstream” of
those carried by NADH
• Transfers electrons to ubiquinone
• Complex III (cytochrome bc1 complex)
• Accepts electrons from ubiquinone from Complex I or II
• 4 protons pumped; electrons transferred to cytochrome c
• Complex IV (cytochrome c oxidase complex)
• Accepts electrons from cytochrome c, pumps 2 protons
• Terminal oxidoreductase, meaning transfers electrons to
terminal electron acceptor (O2)
41.
The Electron Transport Chain of Mitochondria42.
The Electron Transport Chain—Generating ProtonMotive Force
Electron Transport Chain of Prokaryotes
• Tremendous variation: even single species can have
several alternate carriers
• E. coli serves as example of versatility of prokaryotes
• Aerobic respiration in E. coli
• Can use 2 different NADH dehydrogenases
– Proton pump equivalent to complex I of mitochondria
• Succinate dehydrogenase equivalent to complex II of
mitochondria
• Can produce several alternatives to optimally use
different energy sources, including H2
• Lack equivalents of complex III or cytochrome c
– Quinones shuttle electrons directly to ubiquinol
oxidase, a terminal oxidoreductase
– Two versions for high or low O2 concentrations
43.
The Electron Transport Chain—Generating ProtonMotive Force
Electron Transport Chain of Prokaryotes
• Anaerobic respiration in E. coli
• Harvests less energy than aerobic respiration
– Lower electron affinities of terminal electron
acceptors
• Some components different
• Can synthesize terminal oxidoreductase that uses nitrate
as terminal electron acceptor
– Produces nitrite
– E. coli converts to less toxic ammonia
• Sulfate-reducers use sulfate (SO42–) as terminal
electron acceptor
• Produce hydrogen sulfide as end product
44.
The Electron Transport Chain—Generating ProtonMotive Force
45.
The Electron Transport Chain—Generating ProtonMotive Force
ATP Synthase—Harvesting the Proton Motive
Force to Synthesize ATP
• Energy required to establish gradient
• Released when gradient is eased
• ATP synthase allows protons to flow down gradient in
controlled manner
• Uses energy to add phosphate group to ADP
• 1 ATP formed from entry of ~3 protons
• Calculating yields
• Based on experiments on rat mitochondria:
~2.5 ATP made per electron pair from NADH
~1.5 ATP made per electron pair from FADH2
46.
The Electron Transport Chain—Generating ProtonMotive Force
Calculating theoretical maximum yields
• In prokaryotes:
• Glycolysis: 2 NADH 6 ATP
• Transition step: 2 NADH 6 ATP
• TCA Cycle: 6 NADH 18 ATP; 2 FADH2 4 ATP
• Total maximum oxidative phosphorylation yield = 34 ATP
• Slightly less in eukaryotic cells
• NADH from glycolysis in cytoplasm transported across
mitochondrial membrane to enter electron transport chain
– Requires ~1 ATP per NADH generated
47.
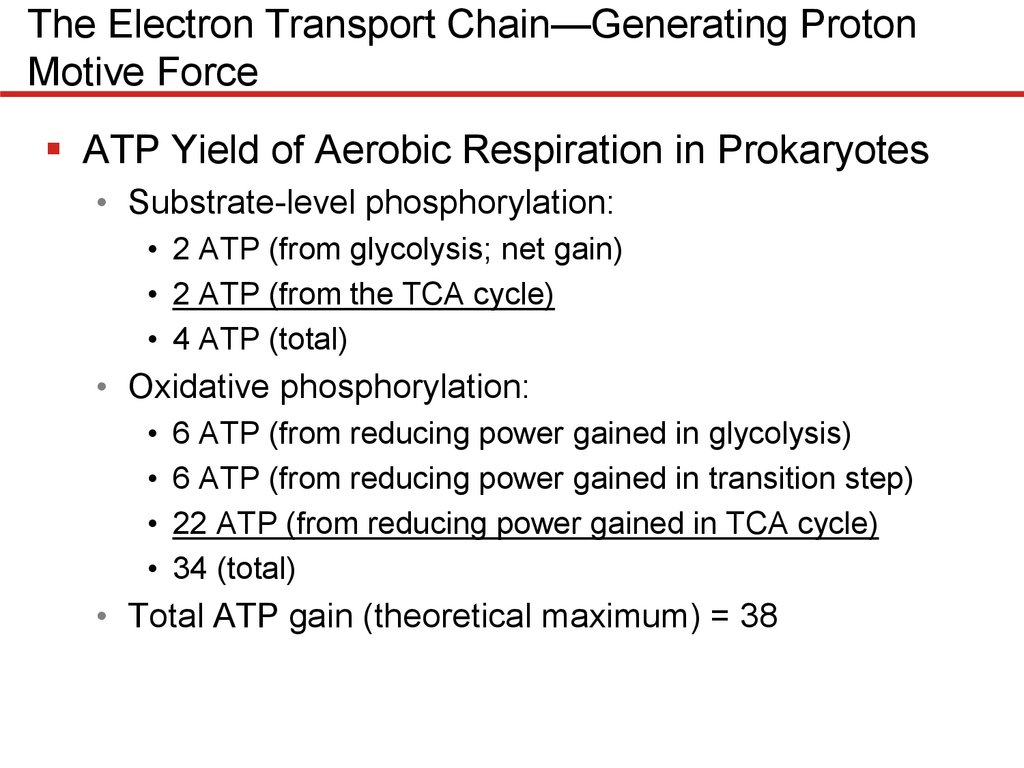
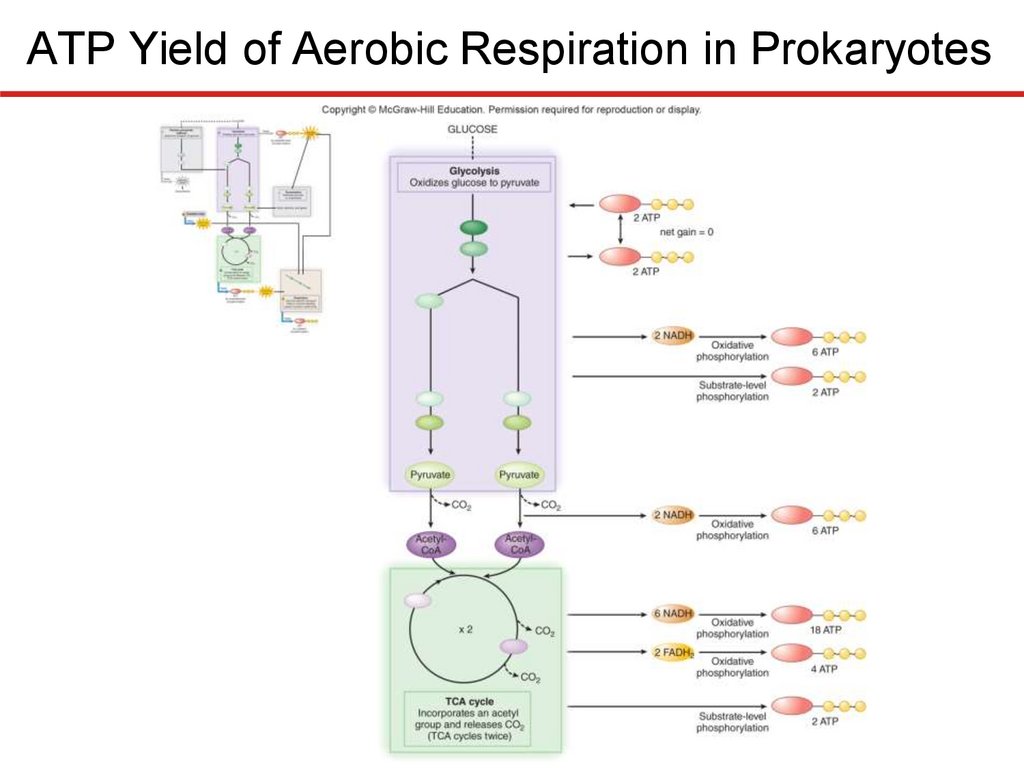
The Electron Transport Chain—Generating ProtonMotive Force
ATP Yield of Aerobic Respiration in Prokaryotes
• Substrate-level phosphorylation:
• 2 ATP (from glycolysis; net gain)
• 2 ATP (from the TCA cycle)
• 4 ATP (total)
• Oxidative phosphorylation:
• 6 ATP (from reducing power gained in glycolysis)
• 6 ATP (from reducing power gained in transition step)
• 22 ATP (from reducing power gained in TCA cycle)
• 34 (total)
• Total ATP gain (theoretical maximum) = 38
48.
ATP Yield of Aerobic Respiration in Prokaryotes49.
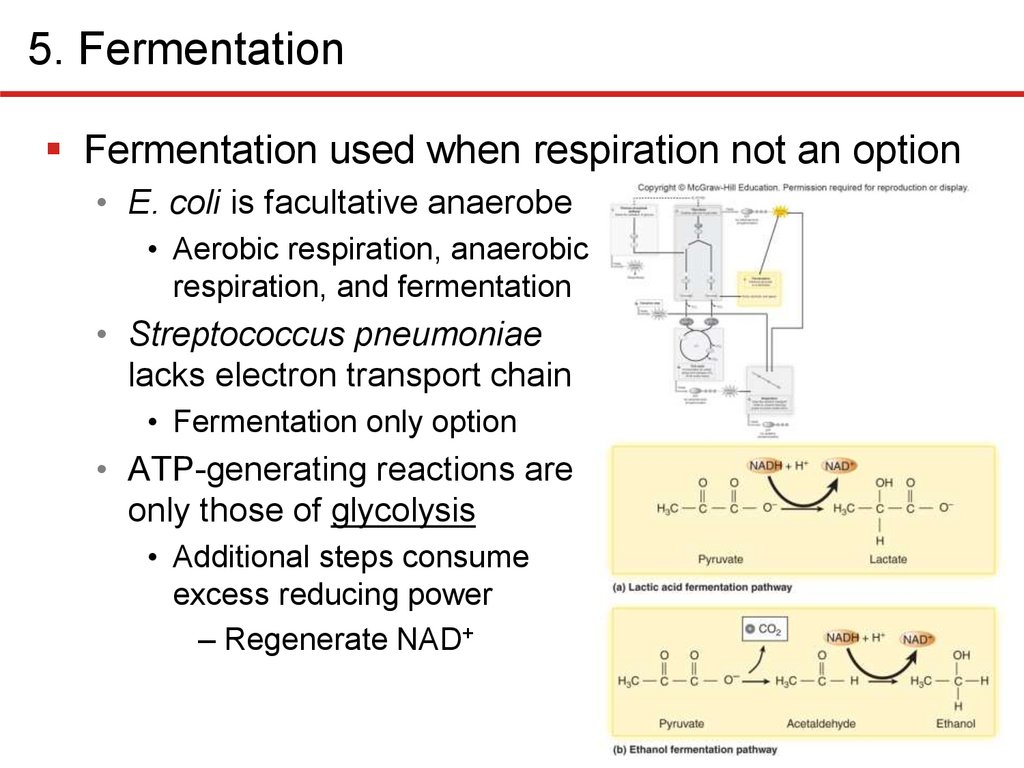
5. FermentationFermentation used when respiration not an option
• E. coli is facultative anaerobe
• Aerobic respiration, anaerobic
respiration, and fermentation
• Streptococcus pneumoniae
lacks electron transport chain
• Fermentation only option
• ATP-generating reactions are
only those of glycolysis
• Additional steps consume
excess reducing power
– Regenerate NAD+
50.
FermentationFermentation end products varied; helpful in
identification, commercially useful
• Ethanol
• Butyric acid
• Propionic acid
• 2,3-Butanediol
• Mixed acids
51.
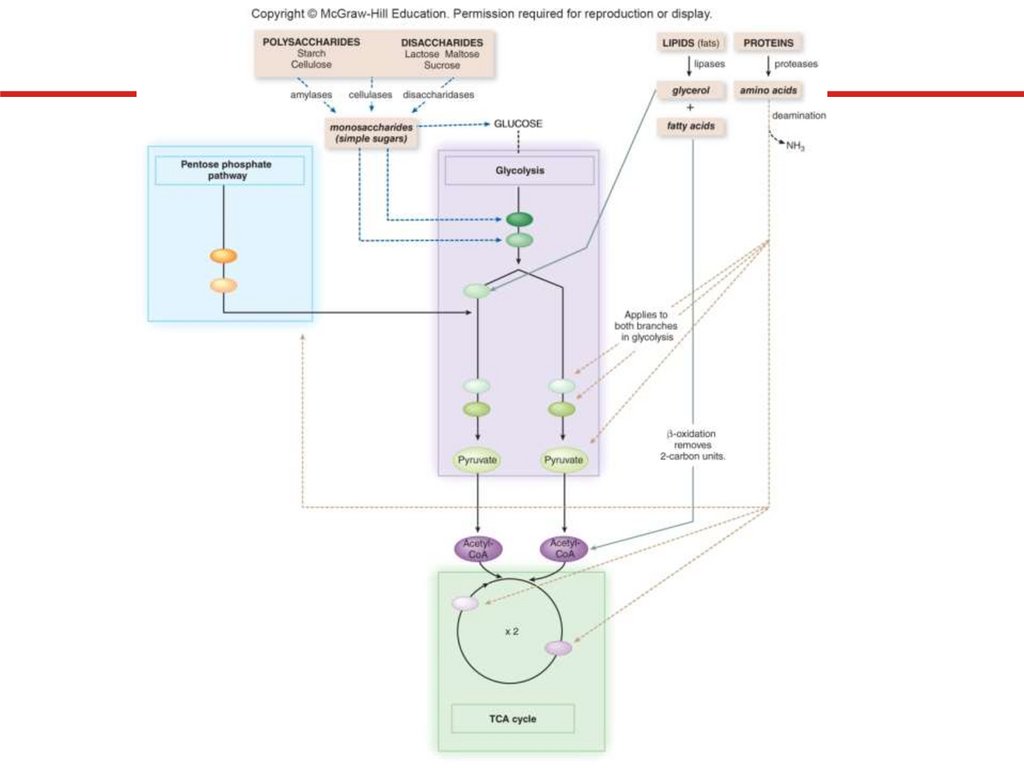
6. Catabolism of Organic Compounds Other thanGlucose
Microbes can use variety of compounds
• Excrete hydrolytic enzymes; transport subunits into cell
• Degrade further to appropriate precursor metabolites
• Polysaccharides and disaccharides
• Amylases digest starch; cellulases digest cellulose
• Disaccharides hydrolyzed by specific disaccharidases
• Lipids
• Fats hydrolyzed by lipases; glycerol converted to
dihydroxyacetone phosphate, enters glycolysis
• Fatty acids degraded by β-oxidation to enter TCA cycle
• Proteins
• Hydrolyzed by proteases; amino group deaminated
• Carbon skeletons converted into precursor molecules
52.
Catabolism of Organic Compounds Other thanGlucose
Microbes can use variety
of compounds
• Convert to precursor
metabolites
• Enter appropriate
metabolic pathways
53.
54.
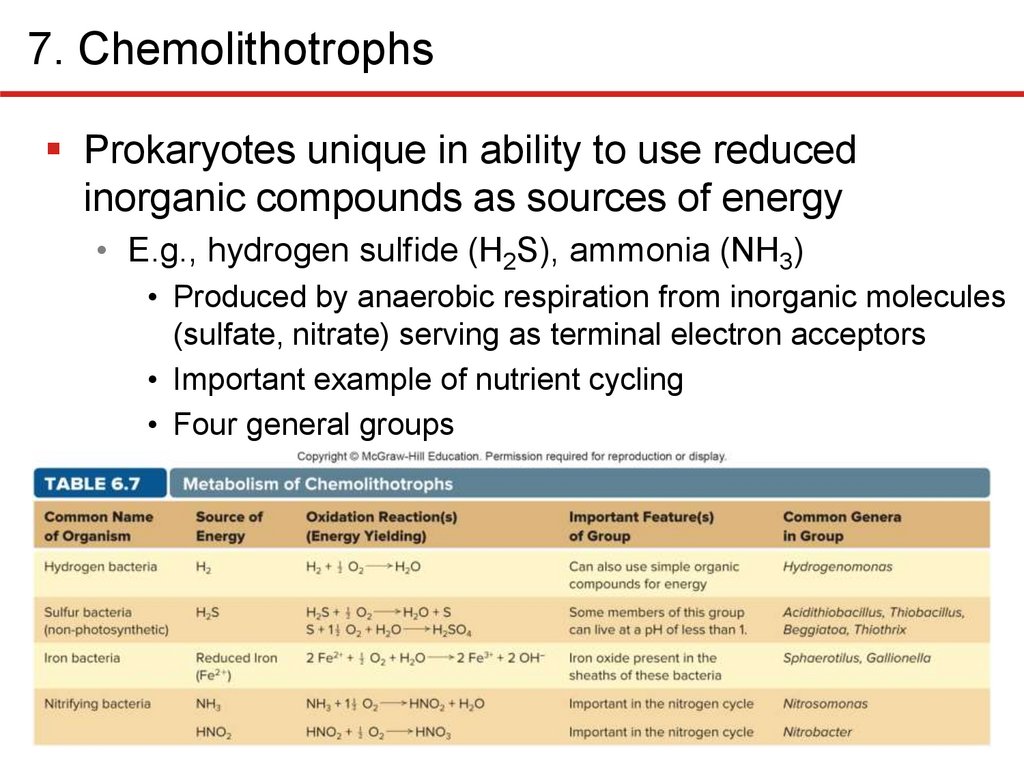
7. ChemolithotrophsProkaryotes unique in ability to use reduced
inorganic compounds as sources of energy
• E.g., hydrogen sulfide (H2S), ammonia (NH3)
• Produced by anaerobic respiration from inorganic molecules
(sulfate, nitrate) serving as terminal electron acceptors
• Important example of nutrient cycling
• Four general groups
55.
8. PhotosynthesisPhotosynthesis
• Plants, algae, several groups of bacteria
• General reaction is
Light Energy
6 CO2 + 12 H2X
C6H12O6 + 12 X + 6 H2O
where X indicates element such as oxygen or sulfur
• Can be considered in two distinct stages
• Light reactions (light-dependent reactions)
– Capture energy and convert it to ATP
• Light-independent reactions (dark reactions)
– Use ATP to synthesize organic compounds
– Involves carbon fixation
56.
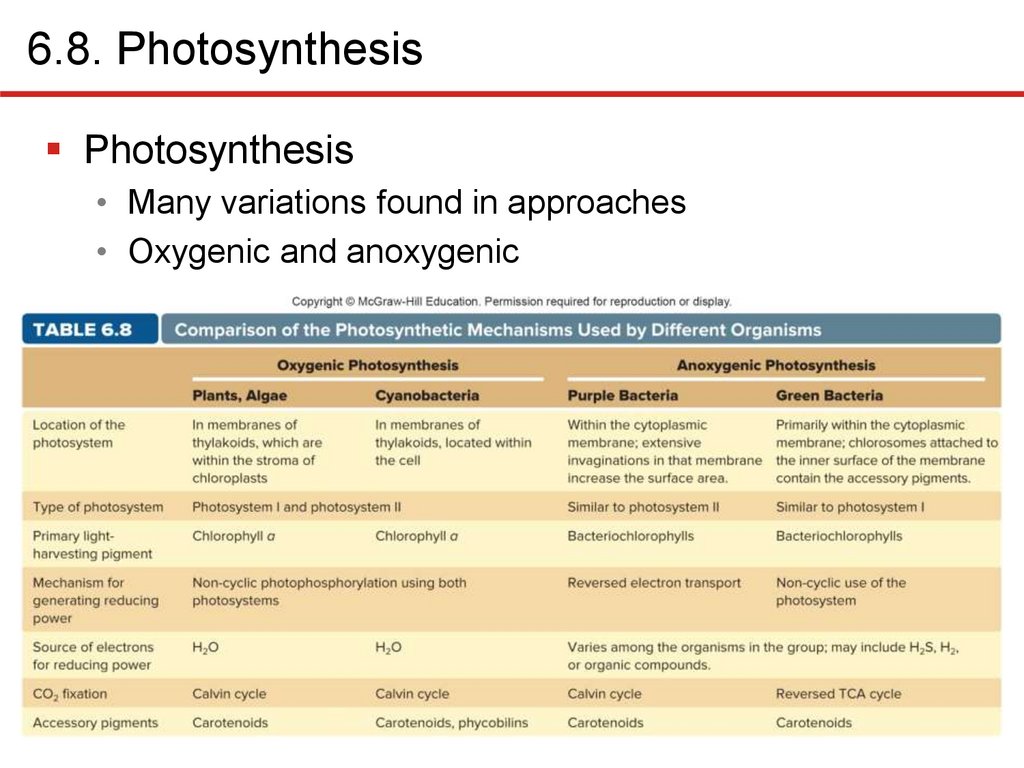
6.8. PhotosynthesisPhotosynthesis
• Many variations found in approaches
• Oxygenic and anoxygenic
57.
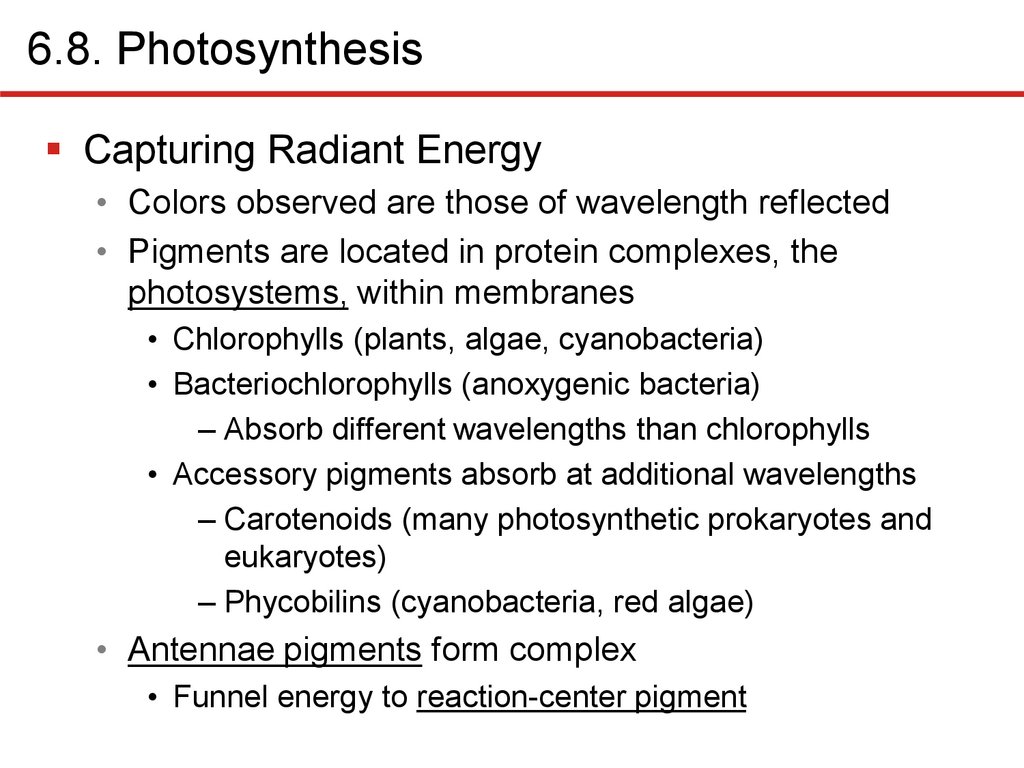
6.8. PhotosynthesisCapturing Radiant Energy
• Colors observed are those of wavelength reflected
• Pigments are located in protein complexes, the
photosystems, within membranes
• Chlorophylls (plants, algae, cyanobacteria)
• Bacteriochlorophylls (anoxygenic bacteria)
– Absorb different wavelengths than chlorophylls
• Accessory pigments absorb at additional wavelengths
– Carotenoids (many photosynthetic prokaryotes and
eukaryotes)
– Phycobilins (cyanobacteria, red algae)
• Antennae pigments form complex
• Funnel energy to reaction-center pigment
58.
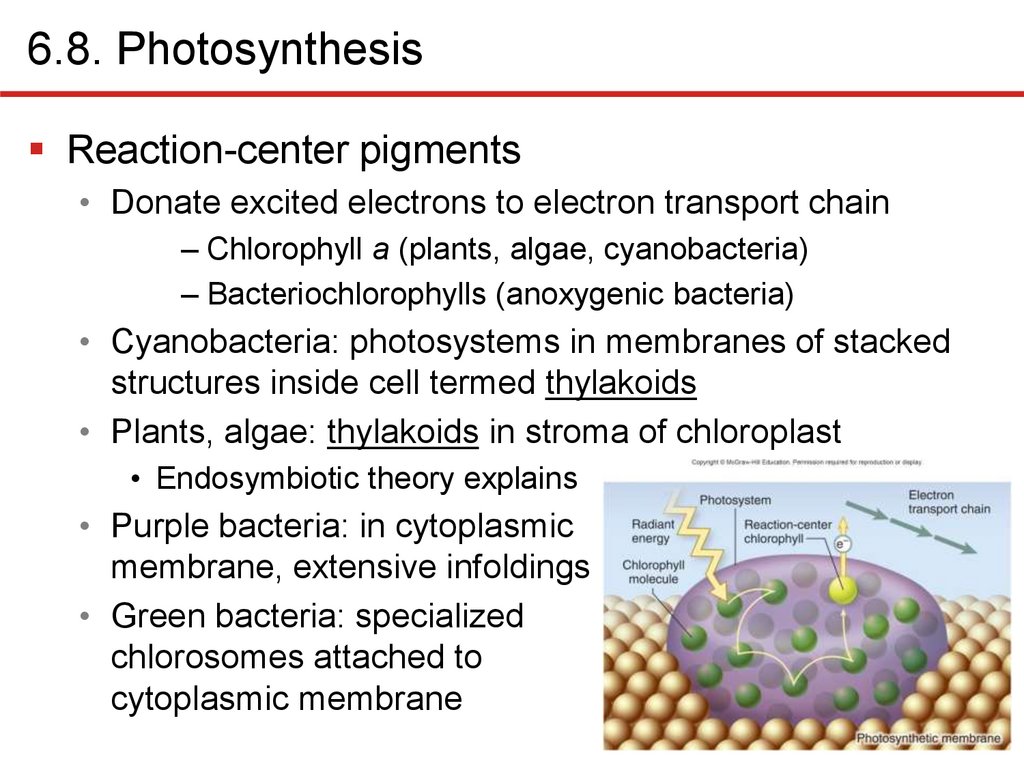
6.8. PhotosynthesisReaction-center pigments
• Donate excited electrons to electron transport chain
– Chlorophyll a (plants, algae, cyanobacteria)
– Bacteriochlorophylls (anoxygenic bacteria)
• Cyanobacteria: photosystems in membranes of stacked
structures inside cell termed thylakoids
• Plants, algae: thylakoids in stroma of chloroplast
• Endosymbiotic theory explains
• Purple bacteria: in cytoplasmic
membrane, extensive infoldings
• Green bacteria: specialized
chlorosomes attached to
cytoplasmic membrane
59.
6.8. Converting Radiant Energy into Chemical EnergyLight-dependent reactions in cyanobacteria and
photosynthetic eukaryotes
• Two distinct photosystems (I and II)
• Cyclic photophosphorylation
– Photosystem I alone produces ATP
– Reaction-center chlorophyll is terminal electron acceptor
• Non-cyclic photophosphorylation
– Used when cells need both ATP and reducing power
• Electrons from photosystem II drive photophosphorylation
– Are then donated to photosystem I
– Photosystem II replenishes electrons by splitting water
– Generates oxygen (process is oxygenic)
• Electrons from photosystem I reduce NADP+ to NADPH
60.
6.8. Converting Radiant Energy into Chemical Energy61.
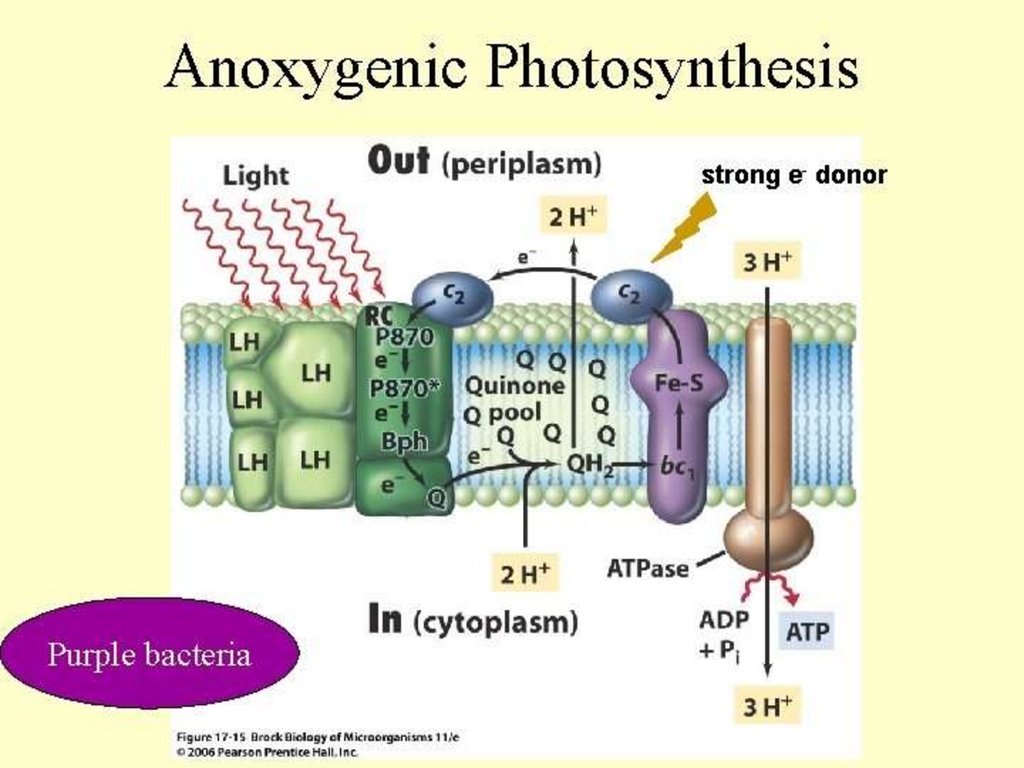
6.8. Converting Radiant Energy into Chemical EnergyLight-dependent reactions in anoxygenic
photosynthetic bacteria
• Each has single photosystem
• Cannot use water as electron donor, so anoxygenic
• Use electron donors such as hydrogen gas (H2), hydrogen
sulfide (H2S), organic compounds
• Purple bacteria: photosystem similar to photosystem II
• Energy of electrons insufficient to reduce NAD+
– Instead expend ATP to use reversed electron transport
• Green bacteria: photosystem similar to photosystem I
• Electrons can generate proton motive force or reduce NAD+
62.
63.
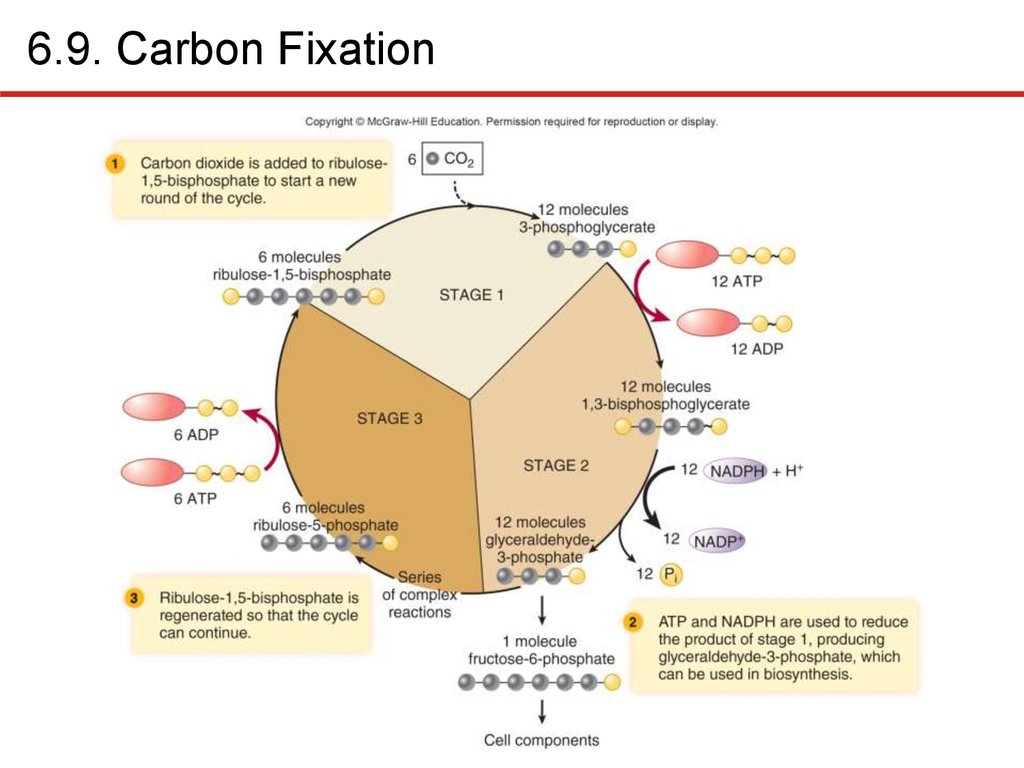
6.9. Carbon FixationChemolithoautotrophs, photoautotrophs use CO2 to
synthesize organic compounds: carbon fixation
• In photosynthetic organisms: light-independent reactions
• Consumes lots of ATP, reducing power
• Reverse process of oxidizing compounds to CO2 liberates a
lot of energy!
• Calvin cycle most commonly used
• Three essential stages
• Incorporation of CO2 into organic compounds
• Reduction of resulting molecule
• Regeneration of starting compound
• Six “turns” of cycle: net gain of one fructose-6-phosphate
• Consumes 18 ATP, 12 NADPH per fructose molecule
64.
6.9. Carbon Fixation65.
6.10. Anabolic Pathways—Synthesizing Subunits fromPrecursor Molecules
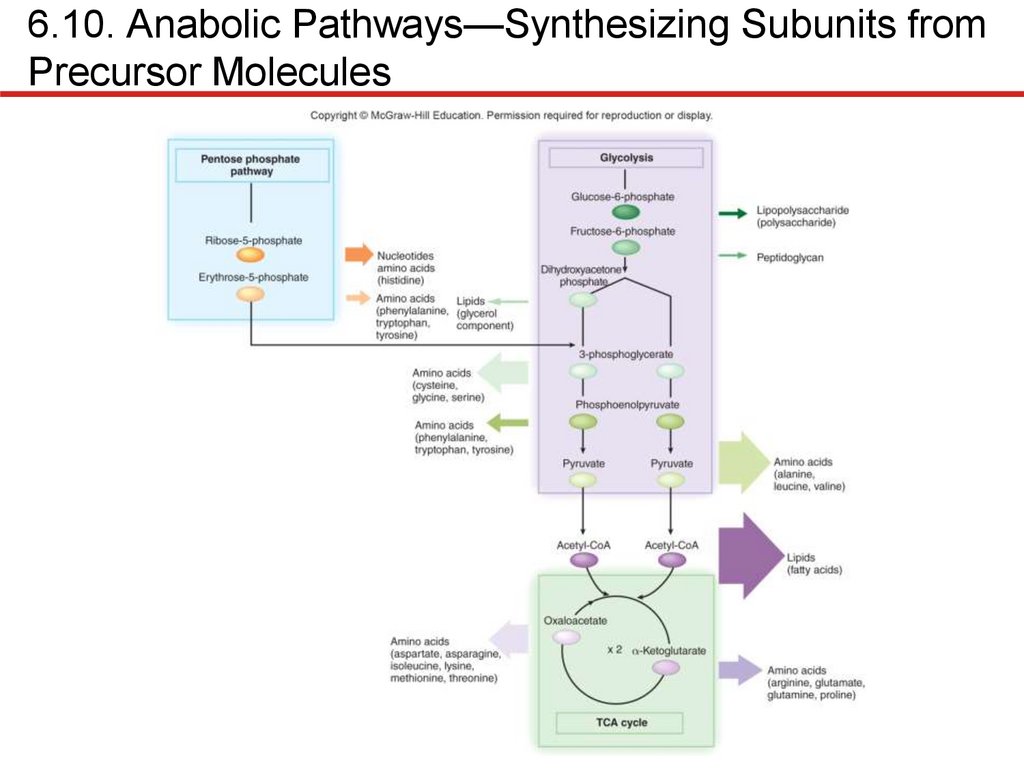
Prokaryotes remarkably similar in biosynthesis
• Synthesize subunits using central metabolic pathways
• If enzymes lacking, end product must be supplied
• Fastidious bacteria require many growth factors
• Lipid synthesis requires fatty acids and glycerol
• Fatty acids: 2-carbon units added to acetyl group from
acetyl-CoA
• Glycerol: dihydroxyacetone phosphate from glycolysis
• Nucleotide synthesis
• DNA, RNA initially synthesized as ribonucleotides
• Purines: atoms added to ribose 5-phosphate to form ring
• Pyrimidines: ring made, then attached to ribose 5-phosphate
• Can be converted to other nucleobases of same type
66.
6.10. Anabolic Pathways—Synthesizing Subunits fromPrecursor Molecules
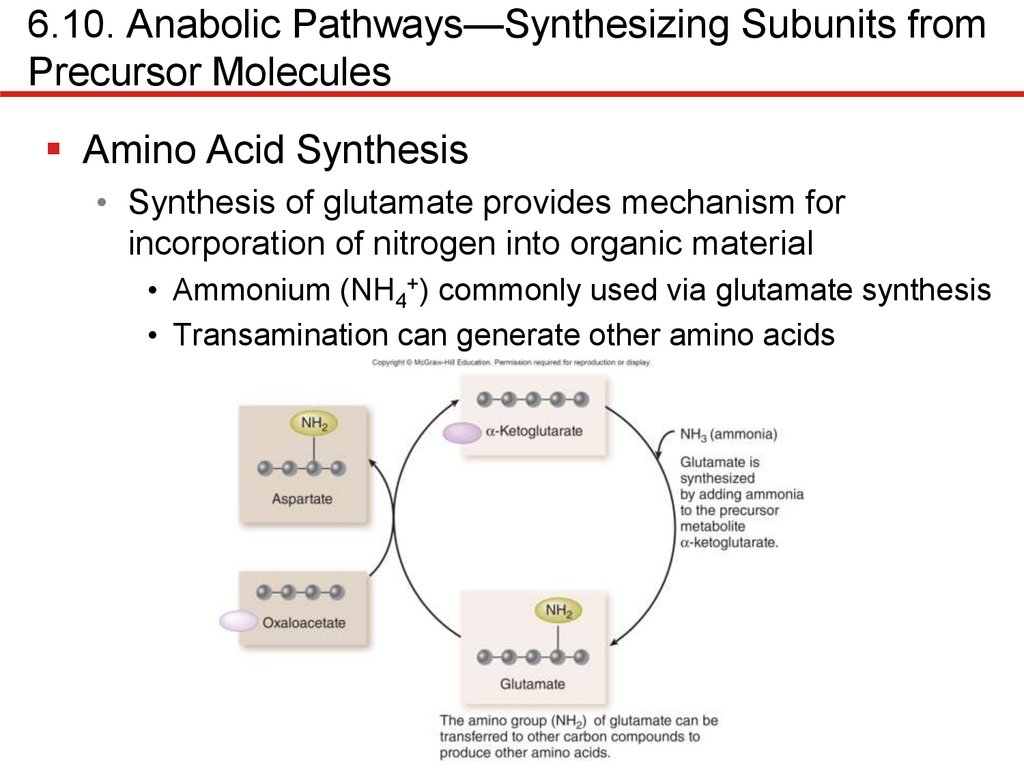
Amino Acid Synthesis
• Synthesis of glutamate provides mechanism for
incorporation of nitrogen into organic material
• Ammonium (NH4+) commonly used via glutamate synthesis
• Transamination can generate other amino acids
67.
6.10. Anabolic Pathways—Synthesizing Subunits fromPrecursor Molecules
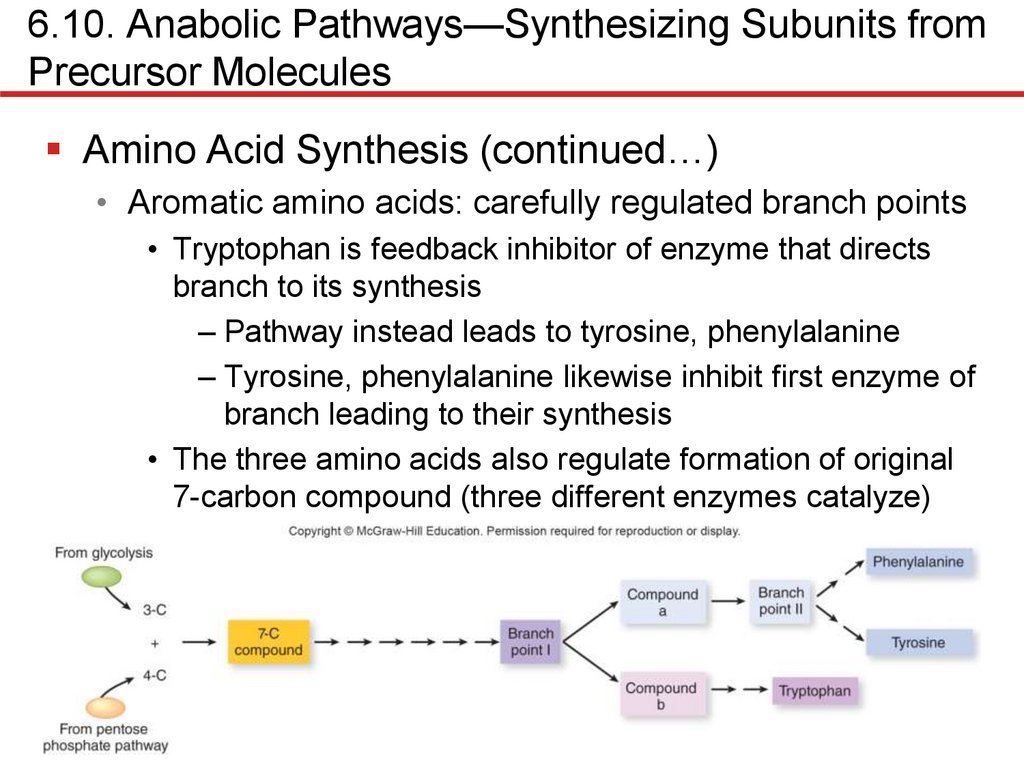
Amino Acid Synthesis (continued…)
• Aromatic amino acids: carefully regulated branch points
• Tryptophan is feedback inhibitor of enzyme that directs
branch to its synthesis
– Pathway instead leads to tyrosine, phenylalanine
– Tyrosine, phenylalanine likewise inhibit first enzyme of
branch leading to their synthesis
• The three amino acids also regulate formation of original
7-carbon compound (three different enzymes catalyze)
68.
6.10. Anabolic Pathways—Synthesizing Subunits fromPrecursor Molecules













 Биология
Биология