Похожие презентации:
UCSC Genome Browser
1.
Школа «Науки о данных»Трек «Биоинформатика» 25-26 апреля
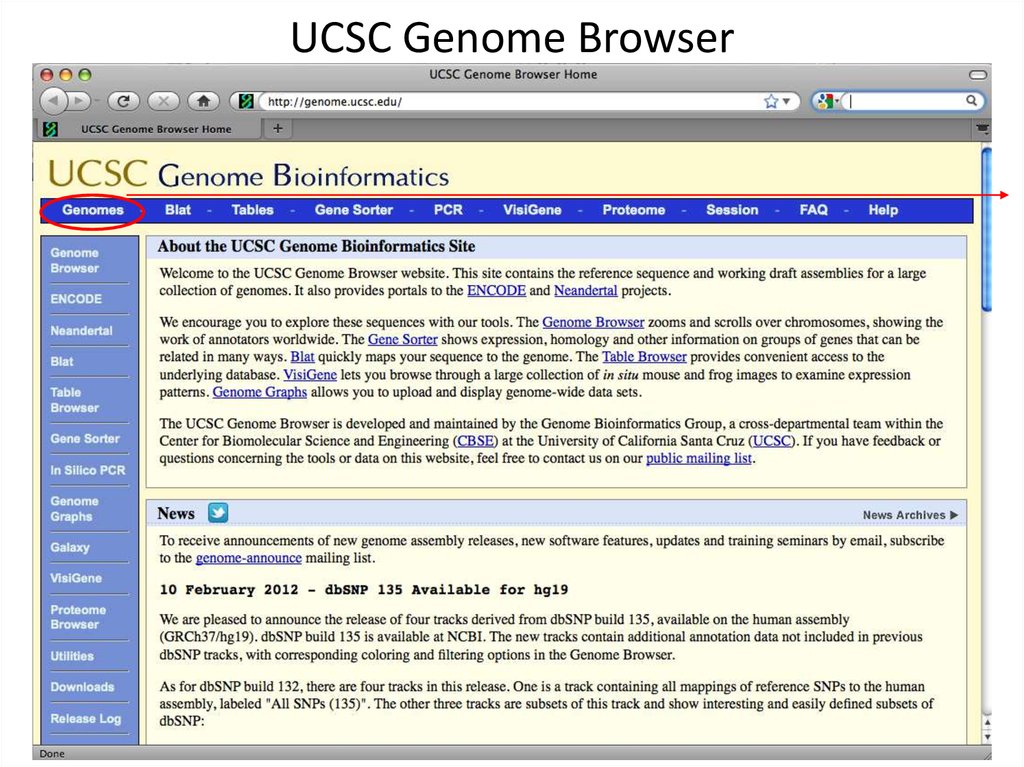
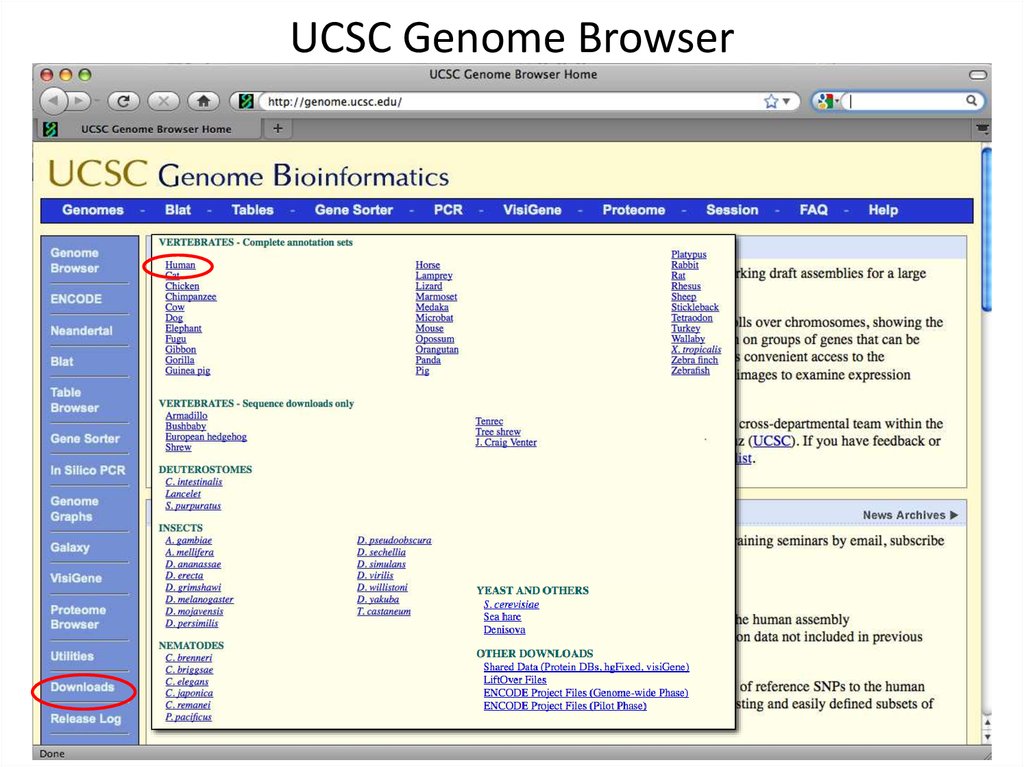
2. UCSC Genome Browser
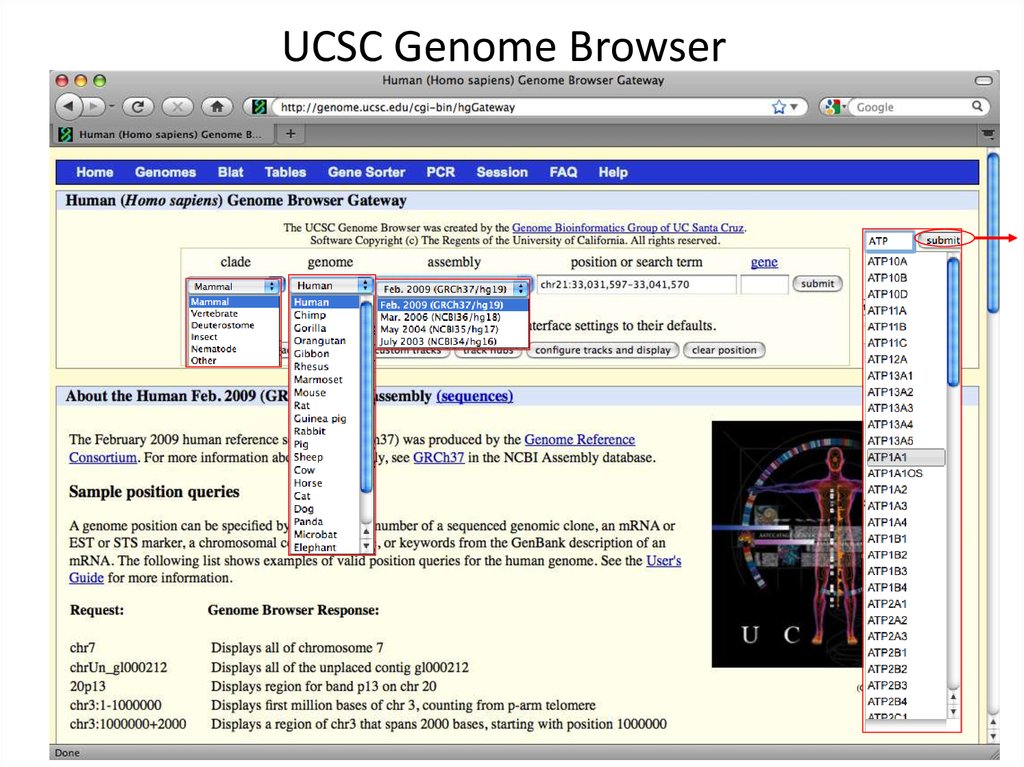
3. UCSC Genome Browser
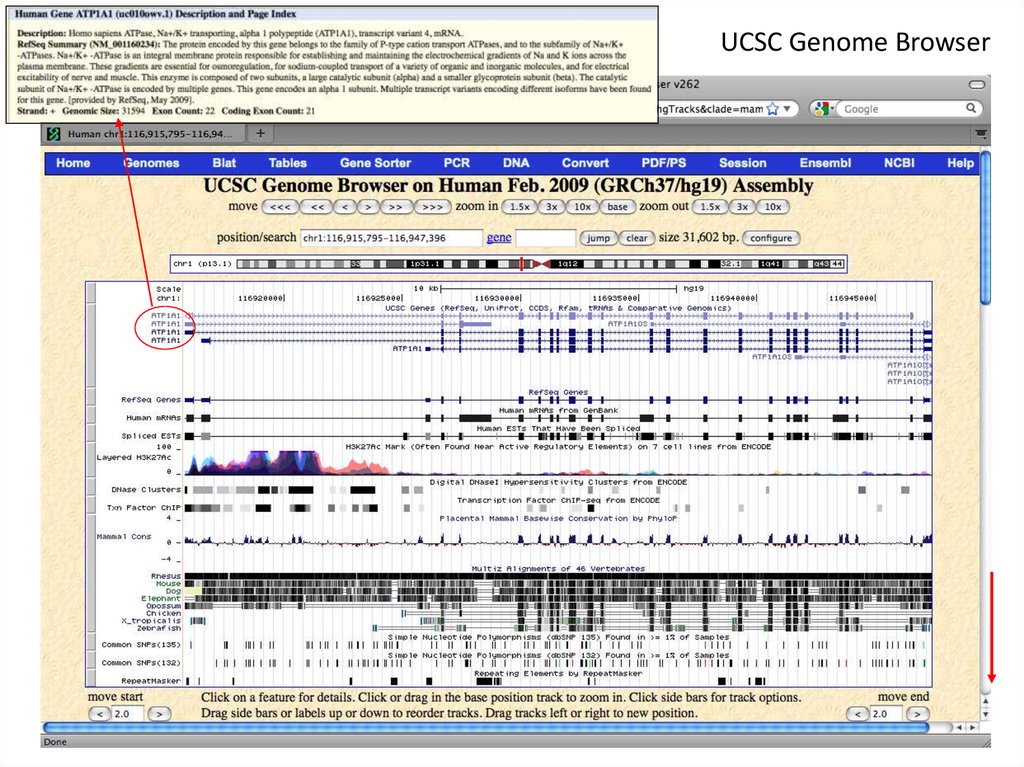
4. UCSC Genome Browser
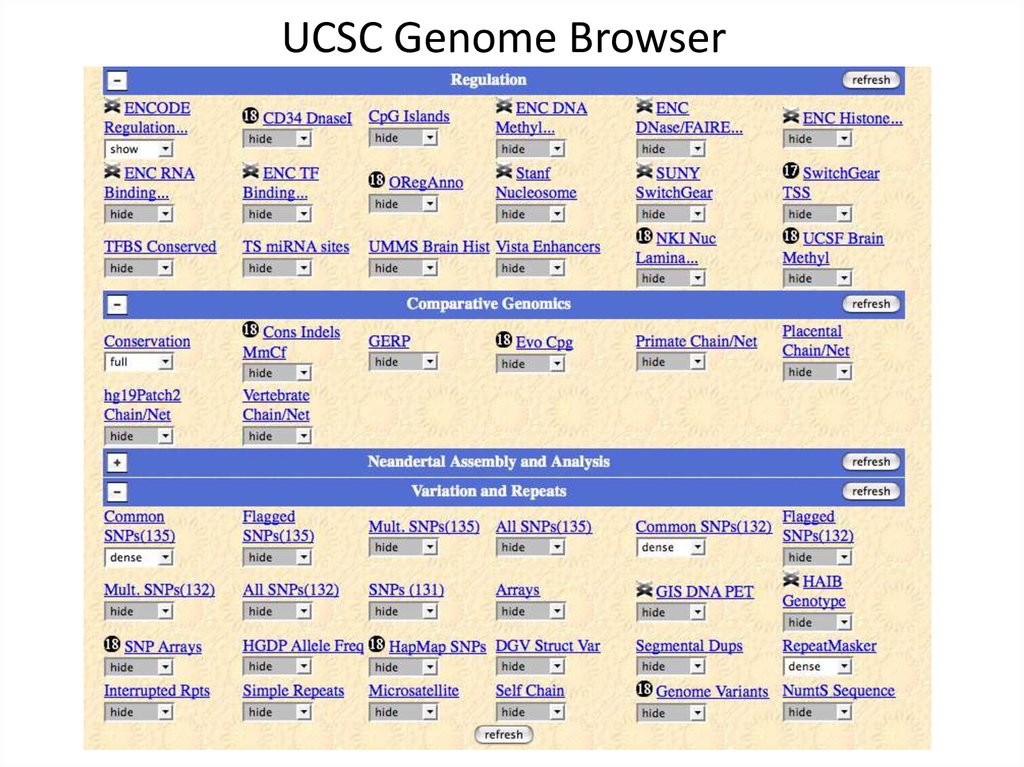
5. UCSC Genome Browser
6. UCSC Genome Browser
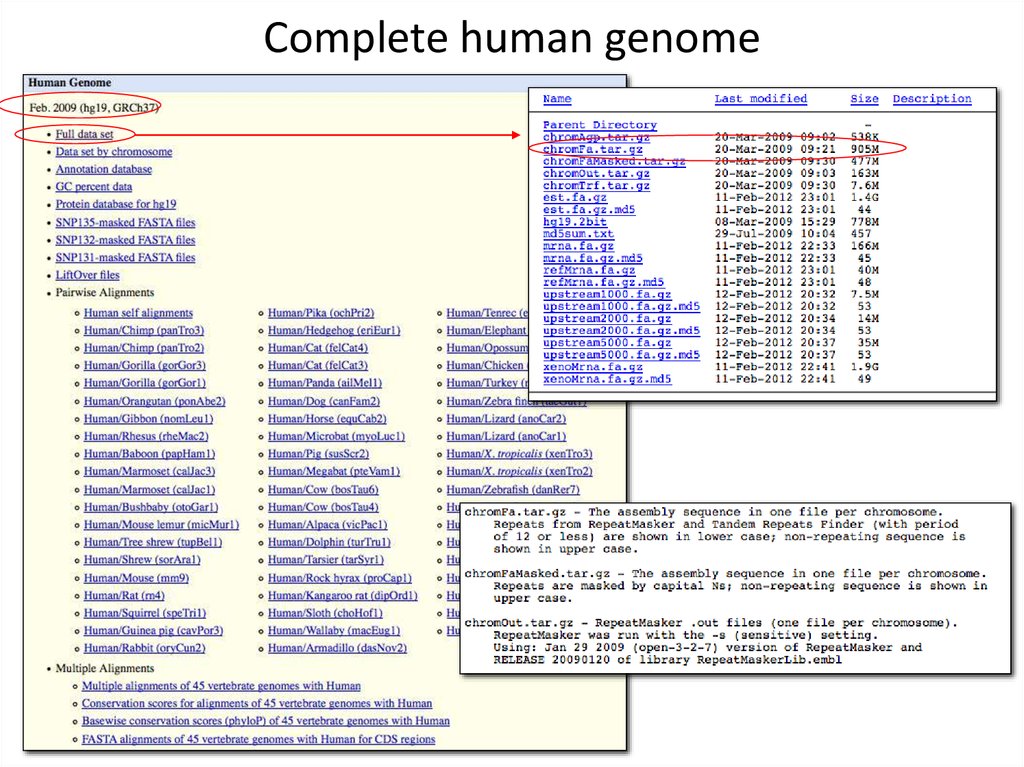
7. Complete human genome
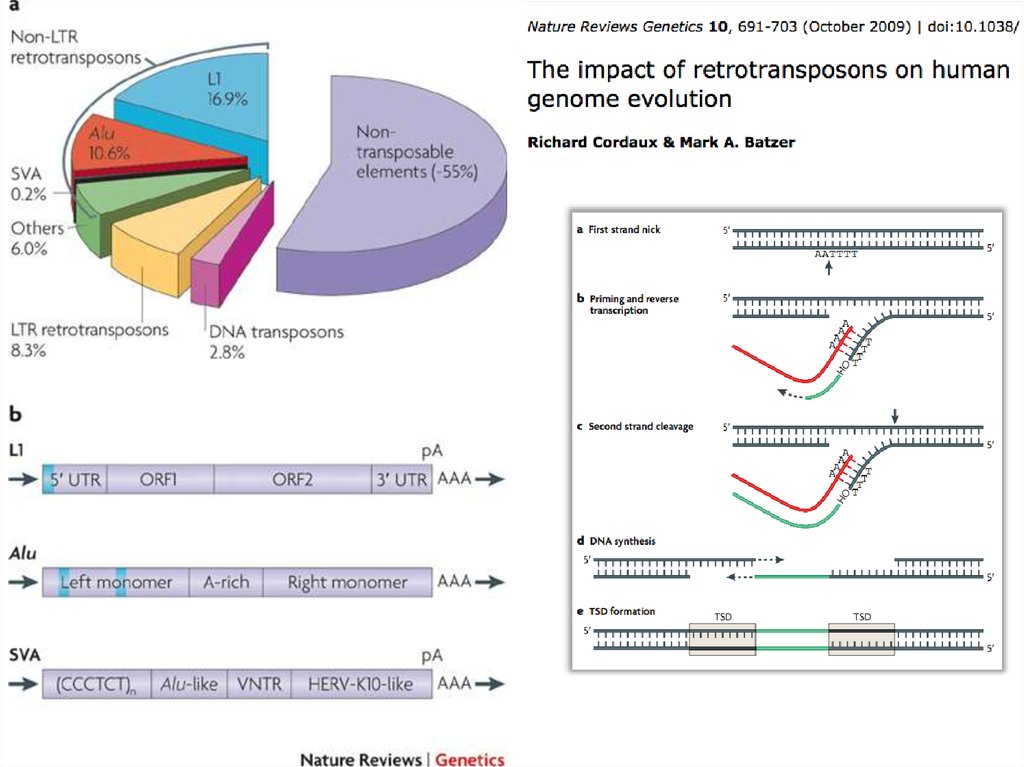
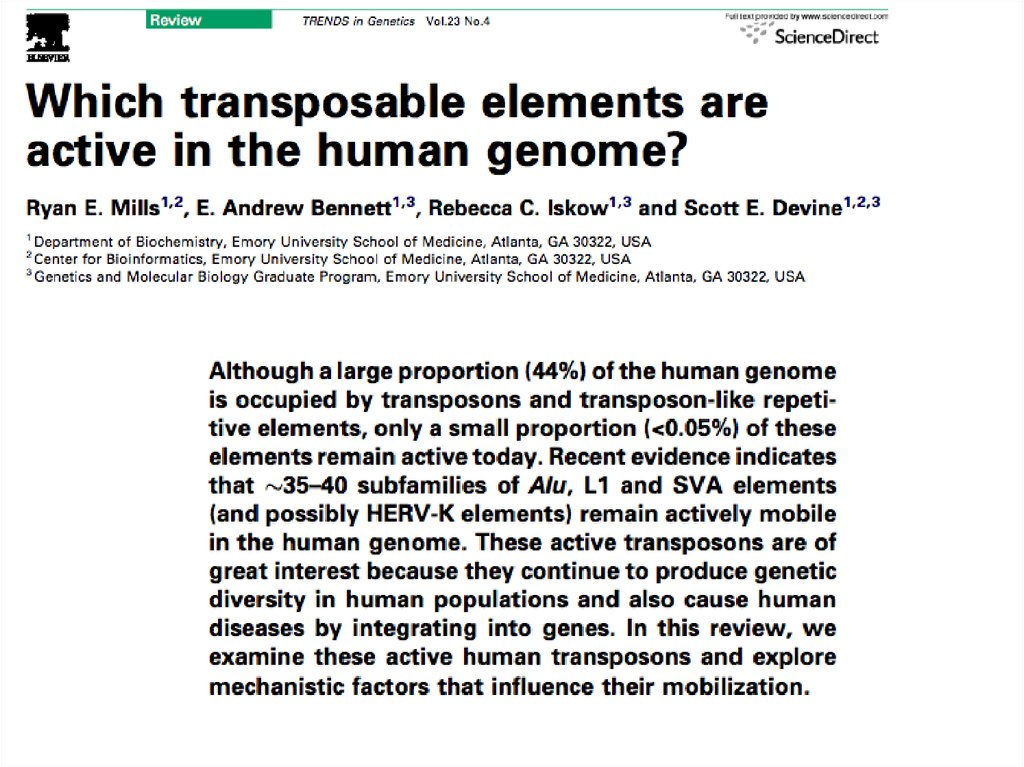
8. Transposable Elements
• 45% of the human genome is occupied bytransposons and transposon-like repetitive
elements.
• Barbara McClintock (1902-1992) in 50s.
• Nobel prize in 1983
9.
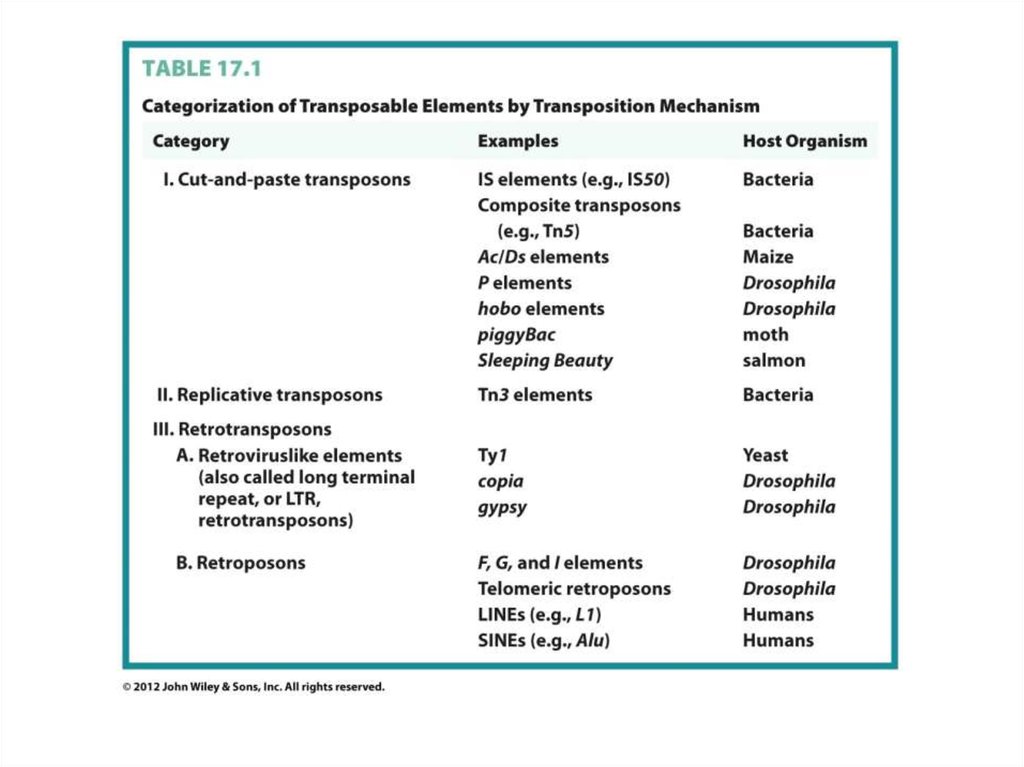
Class IIClass I – retrotransposons (via RNA intermediate)
Схожесть с ретровирусами
Retrovirus reverse transcripiton
http://www.youtube.com/watch?v=eS1GODinO8w
10.
11.
12.
13.
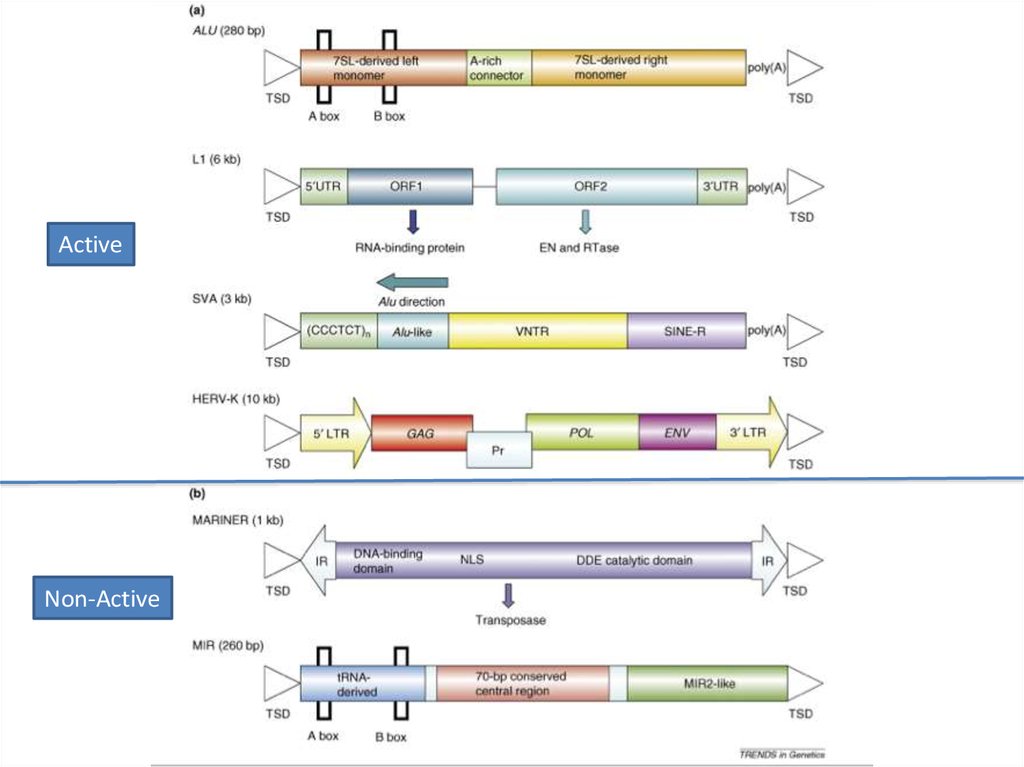
ActiveNon-Active
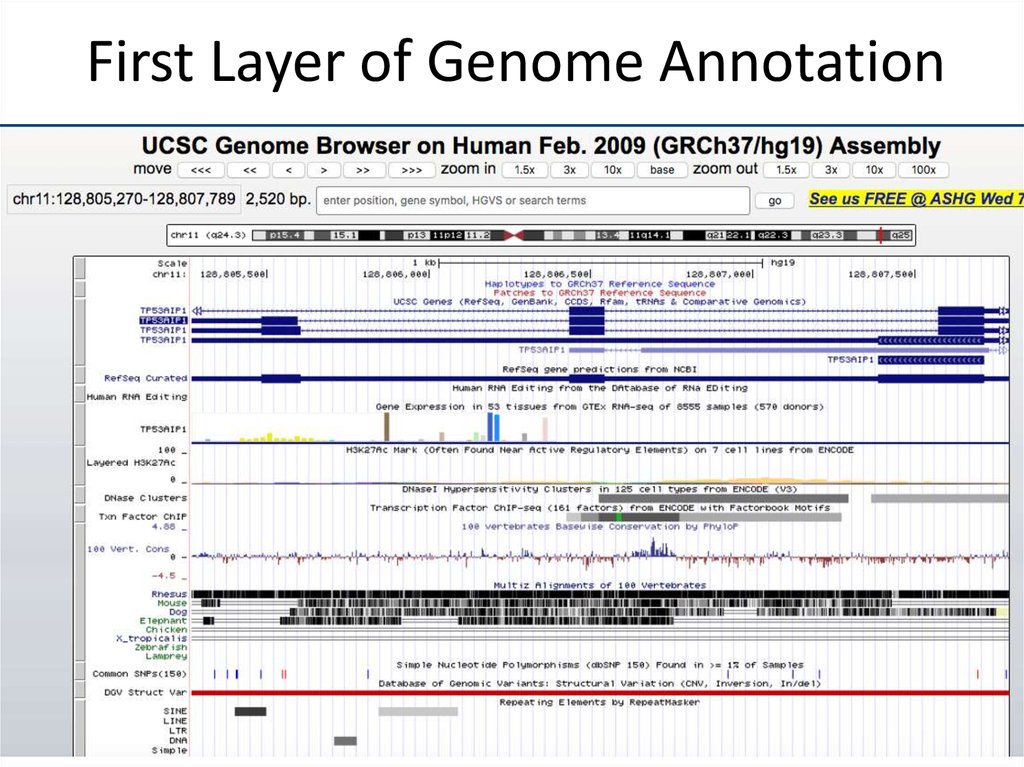
14. First Layer of Genome Annotation
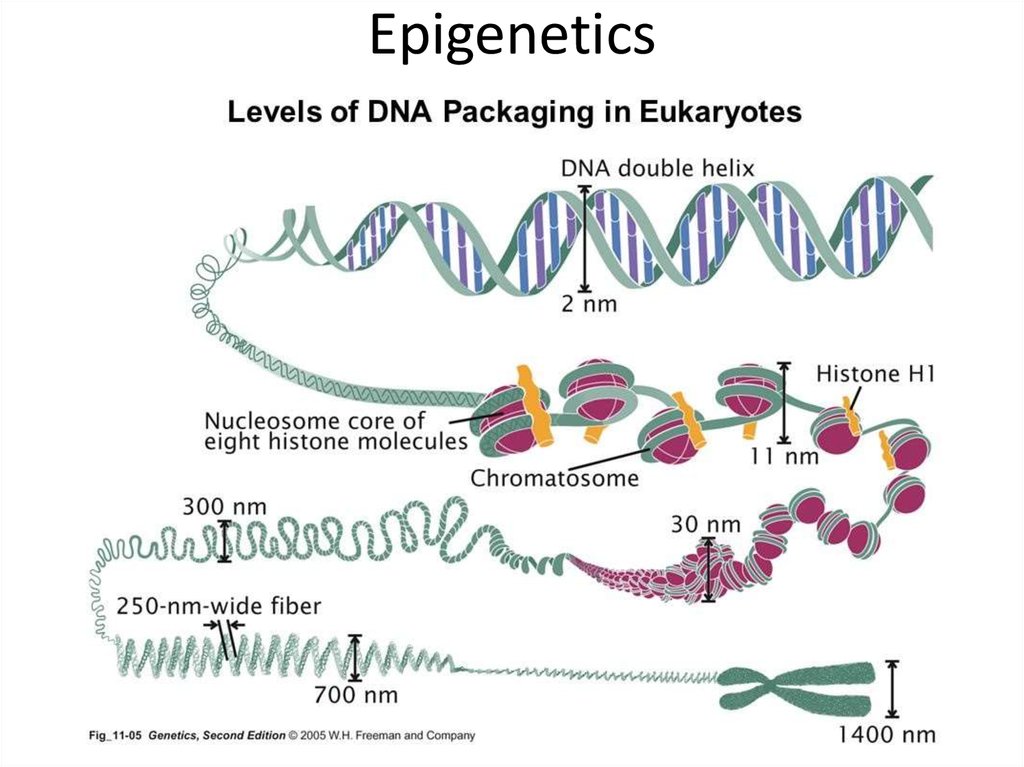
15. Epigenetics
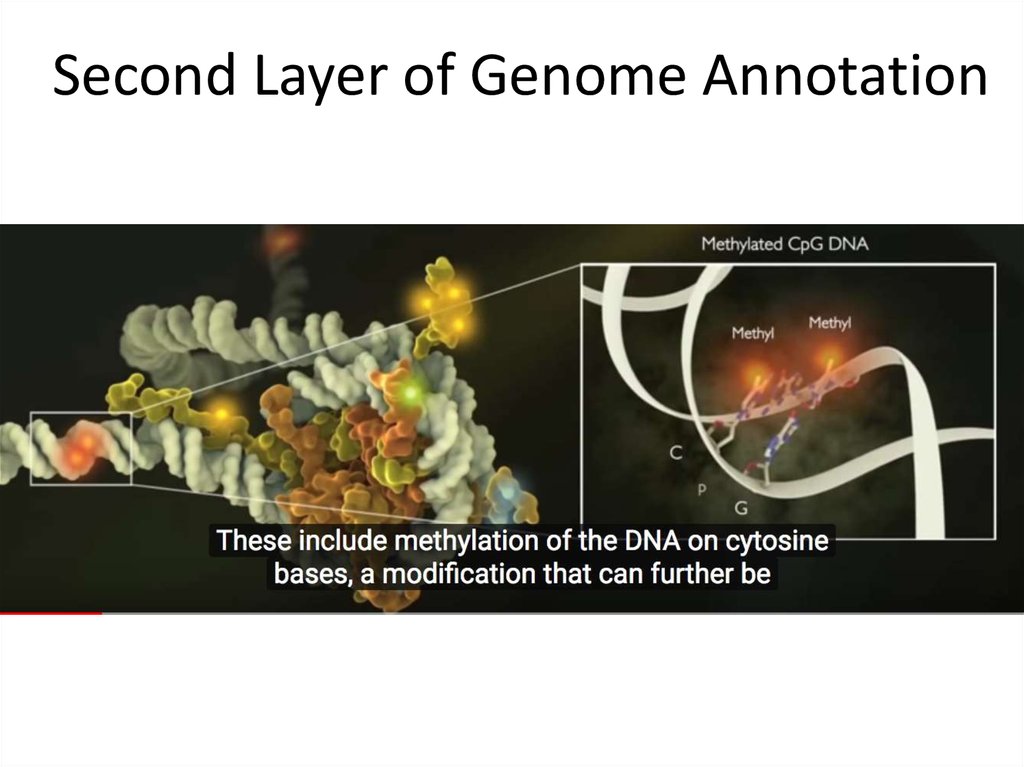
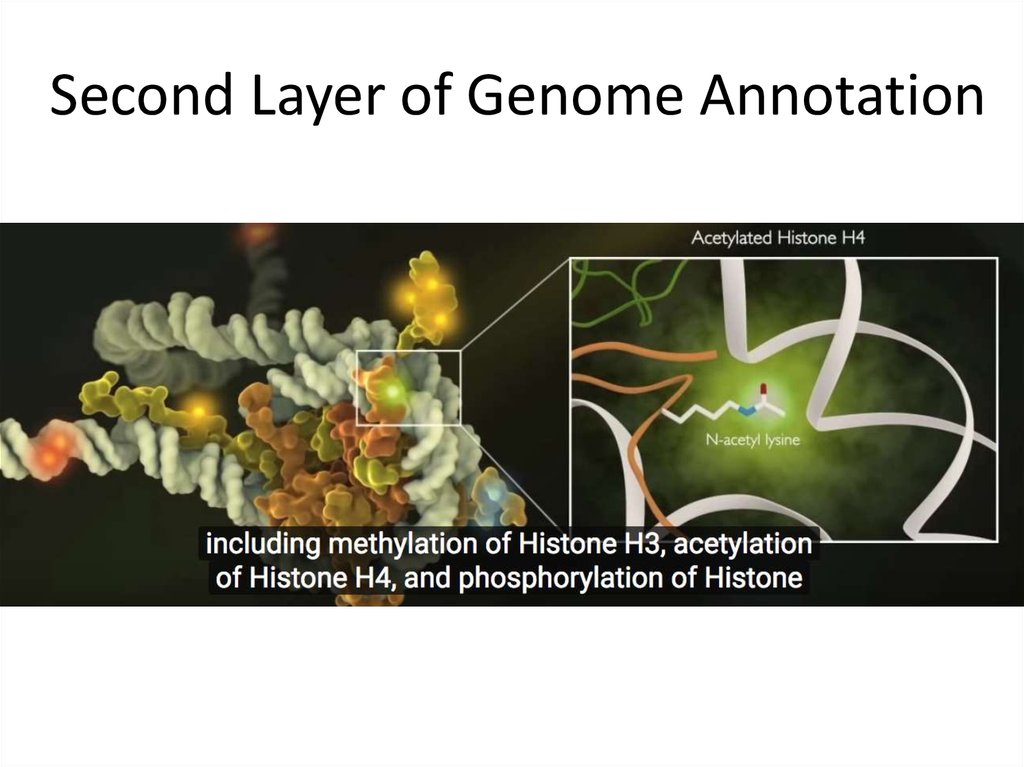
16. Second Layer of Genome Annotation
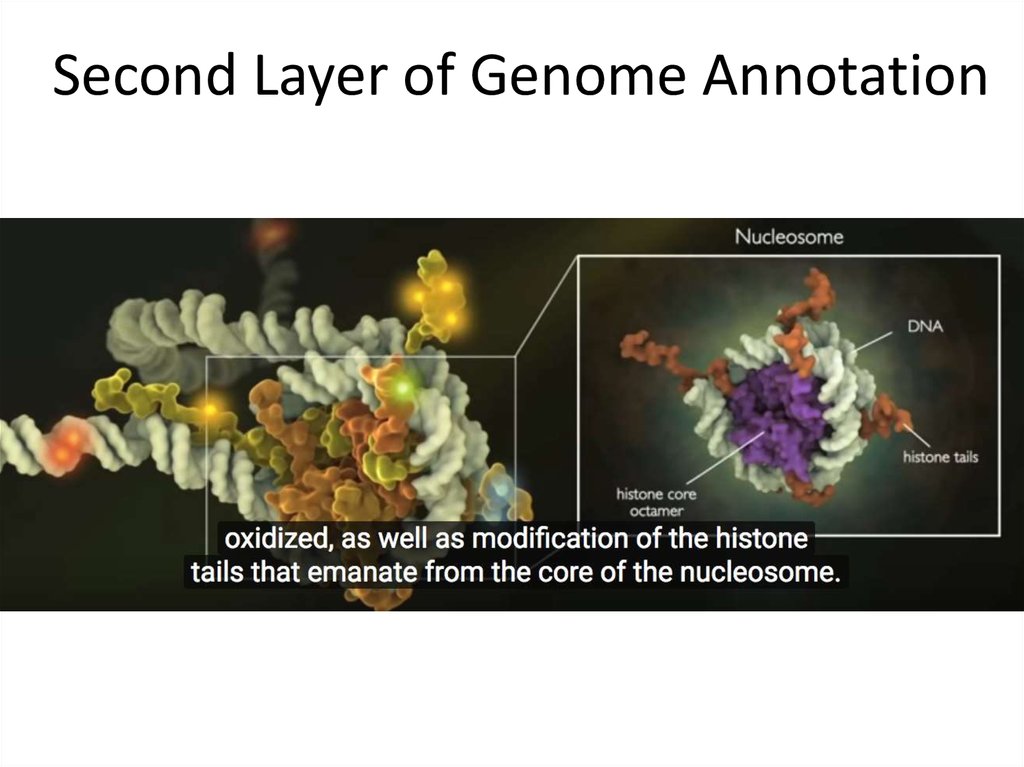
17. Second Layer of Genome Annotation
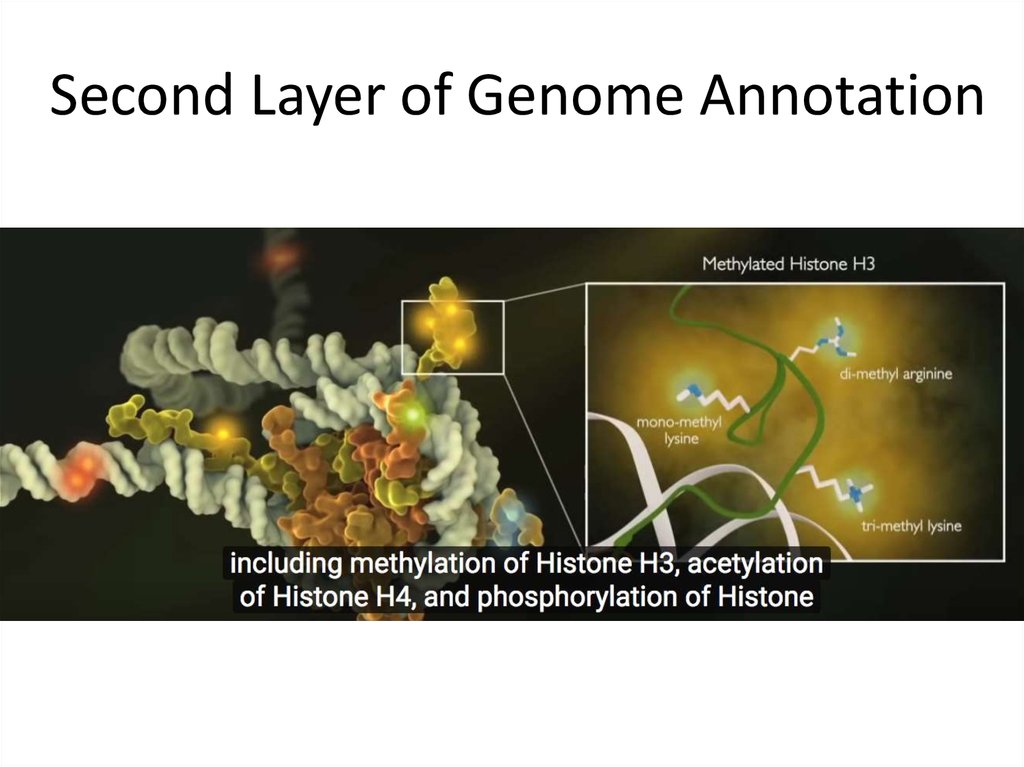
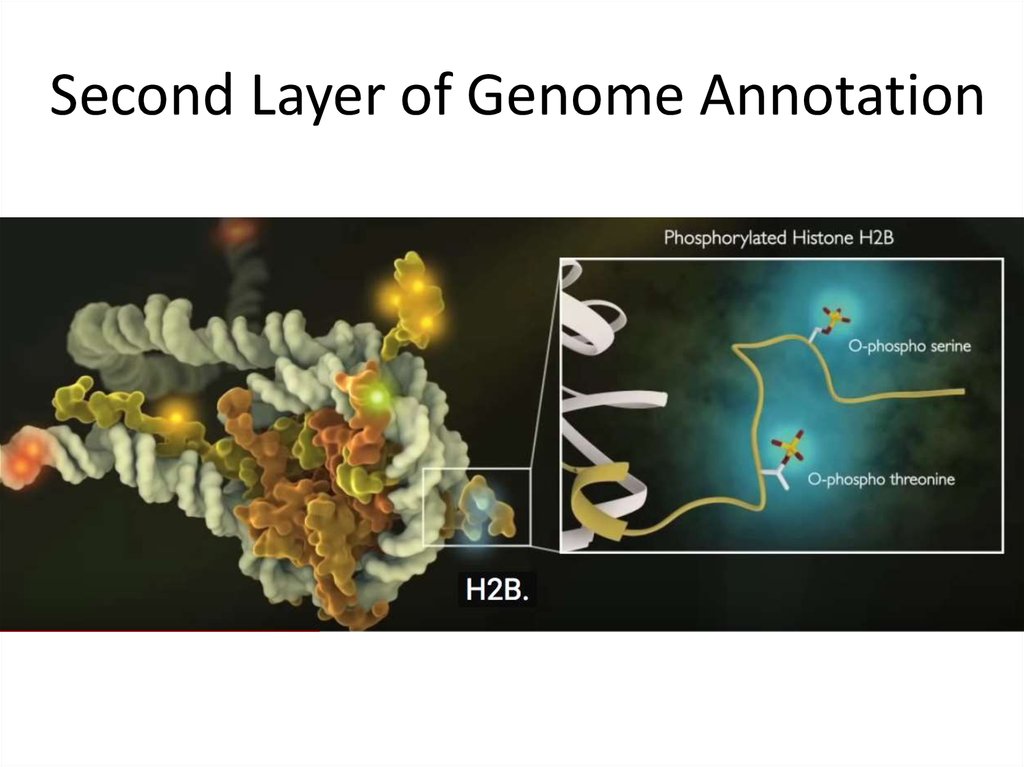
18. Second Layer of Genome Annotation
19. Second Layer of Genome Annotation
20. Second Layer of Genome Annotation
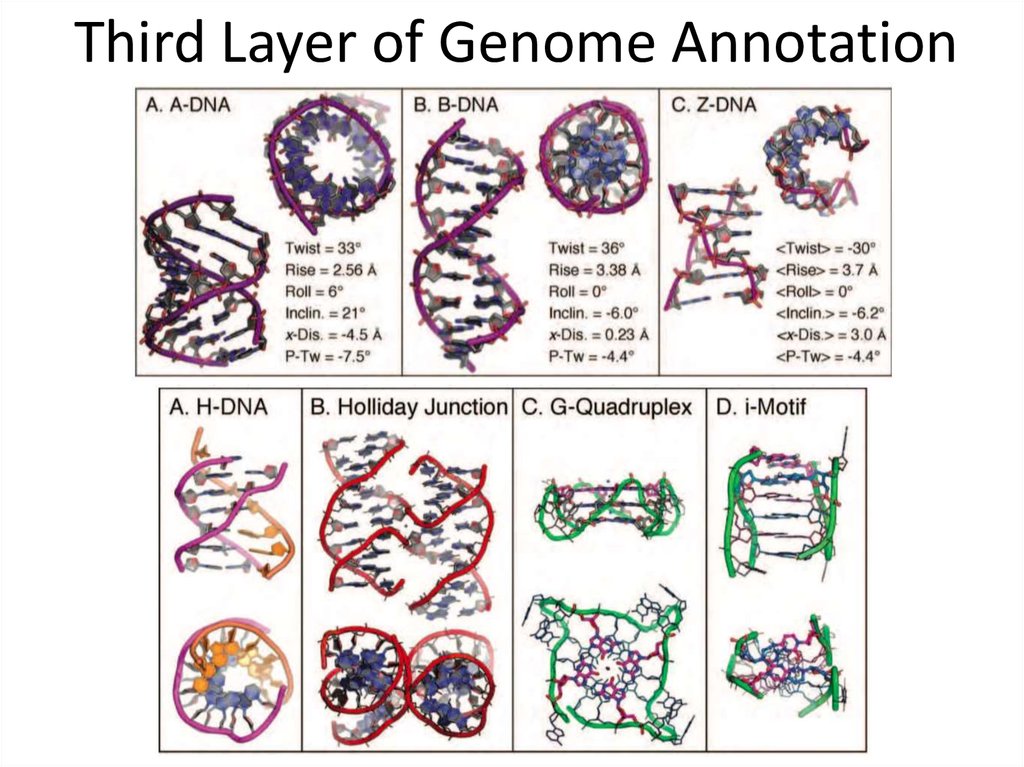
21.
Third Layer of Genome Annotation22. Data Accumulation
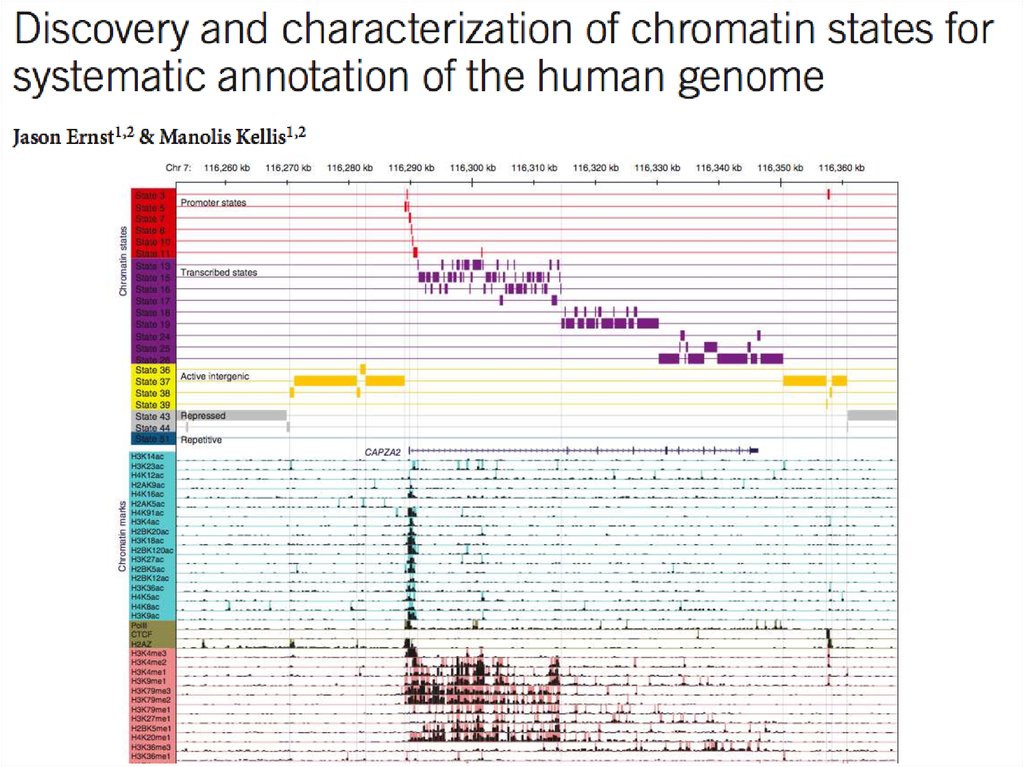
23. ENCODE: Encyclopedia of DNA Elements
24. Digital Universe
• Like the Physical Universe the DigitalUniverse is also expanding but much
faster doubling every two years – and by
2020 will be 44 zettabytes (10^ 21)
• Every second a new 205 000 bytes come
to being
• At the end of this lecture the digital
universe will grow by 2 214 000 000
bytes or 2.2 GB.
25. Digital Universe
Data Universe Will Expand To 44 Trillion GBs By 202026. Что делать?
27. Что получилось? (Success Stories)
28. СКРЫТЫЕ ЦЕПИ МАРКОВА
01
1
1
…
1
2
2
2
…
2
…
…
…
K
K
K
x1
x2
x3
…
…
0
K
xL
H1
H2
Hi
HL-1
HL
X1
X2
Xi
XL-1
XL
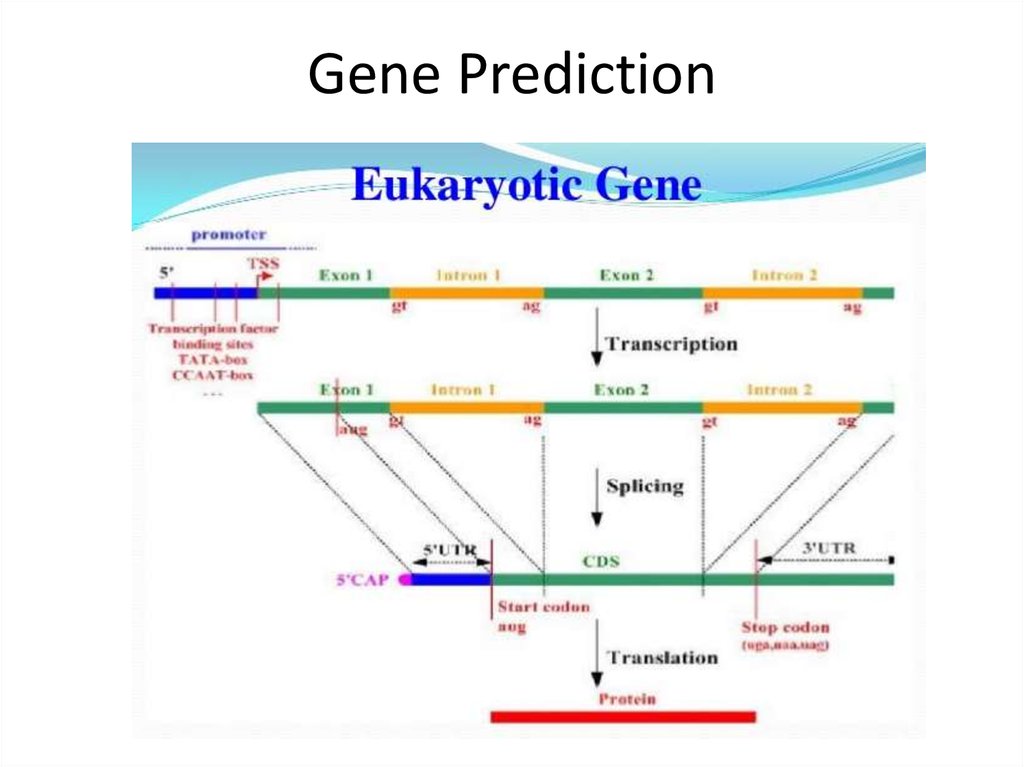
29. Gene Prediction
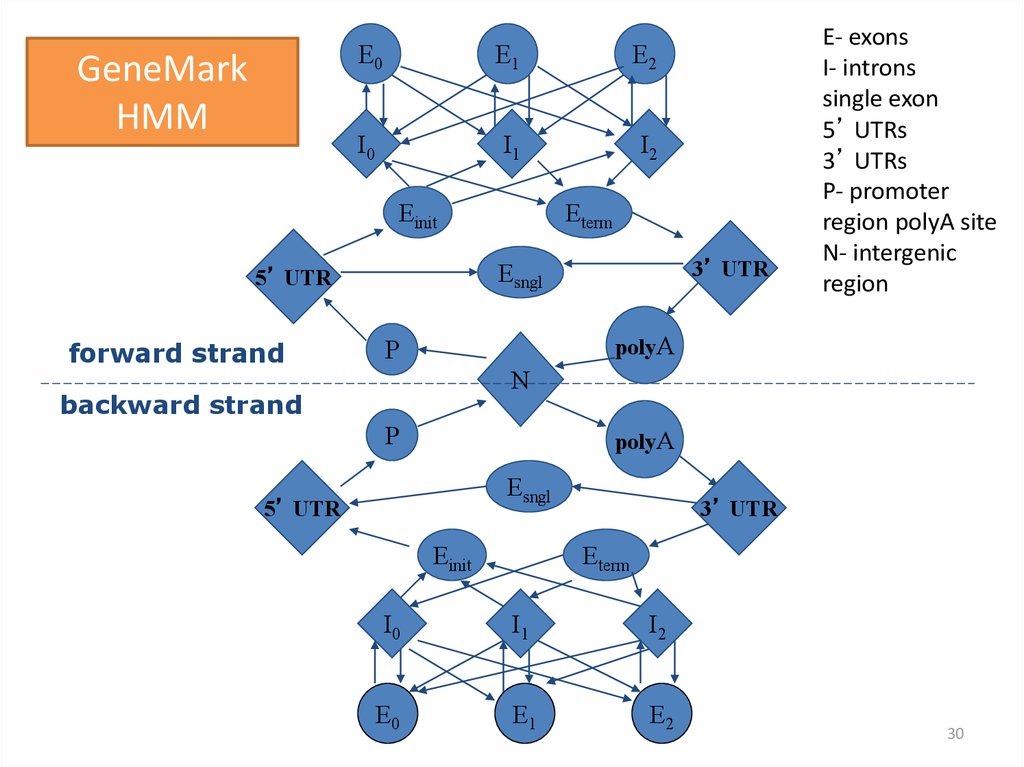
30.
GeneMarkHMM
E0
E1
E2
I0
I1
I2
Einit
3’ UTR
Esngl
5’ UTR
forward strand
Eterm
E- exons
I- introns
single exon
5’ UTRs
3’ UTRs
P- promoter
region polyA site
N- intergenic
region
polyA
P
N
backward strand
P
polyA
Esngl
5’ UTR
Einit
3’ UTR
Eterm
I0
I1
I2
E0
E1
E2
30
31. Promoter prediction
McPromoter• Hidden Markov model with six interpolated
Markov chain submodels
– upstream 1 and 2,
– TATA box, spacer,
– Initiator
– downstream.
– Gaussian densities of DNA physicochemical
properties.
• Neural network classifier
32.
Nature 2010• predict tissue-dependent changes in
alternative splicing for thousands of exons.
• 1,014 features: known motifs, new motifs,
short motifs and features describing transcript
structure
• trained on RNA-seq data
• single-layer logistic Bayesian network or
neural network, or a weighted combination of
single-layer decision trees.
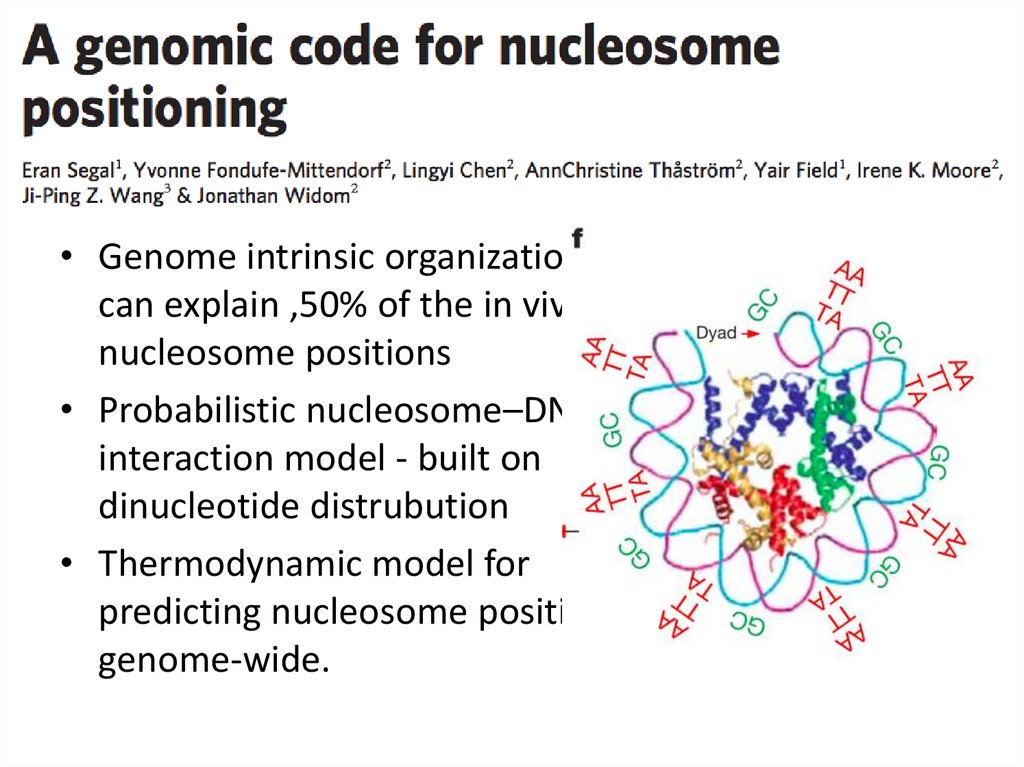
33.
• Genome intrinsic organizationcan explain ,50% of the in vivo
nucleosome positions
• Probabilistic nucleosome–DNA
interaction model - built on
dinucleotide distrubution
• Thermodynamic model for
predicting nucleosome positions
genome-wide.
34.
35.
Schematic overview of epigenetic regulatory mechanisms.Yonggang Zhou et al. Circ Res. 2011;109:1067-1081
Copyright © American Heart Association, Inc. All rights reserved.
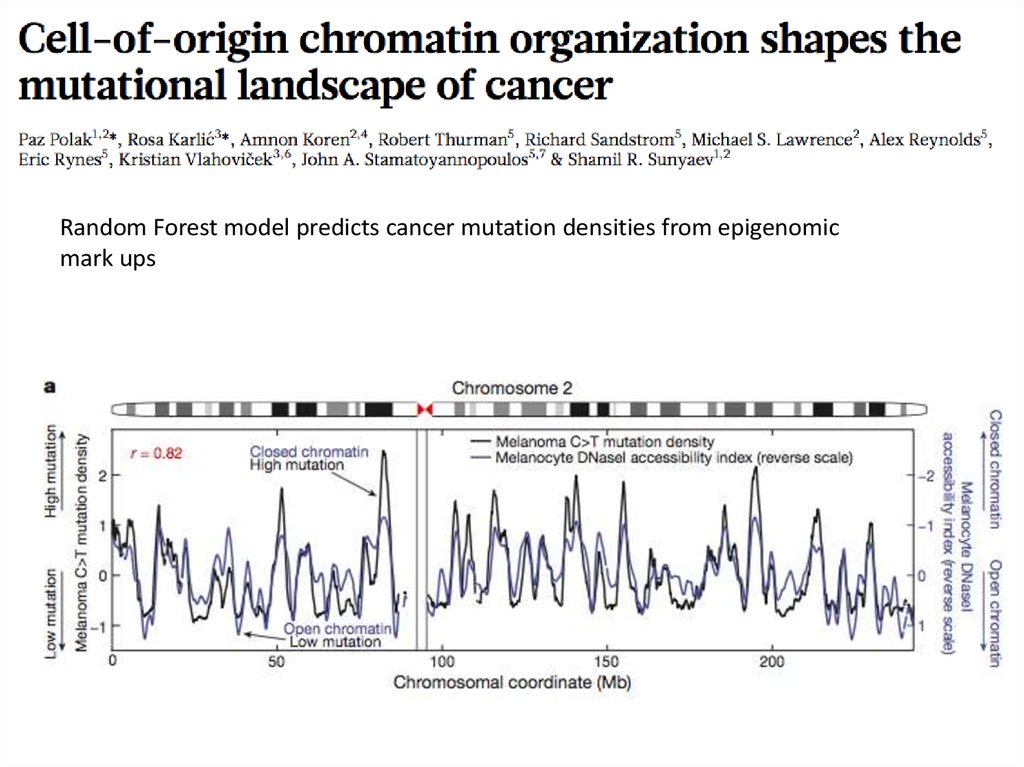
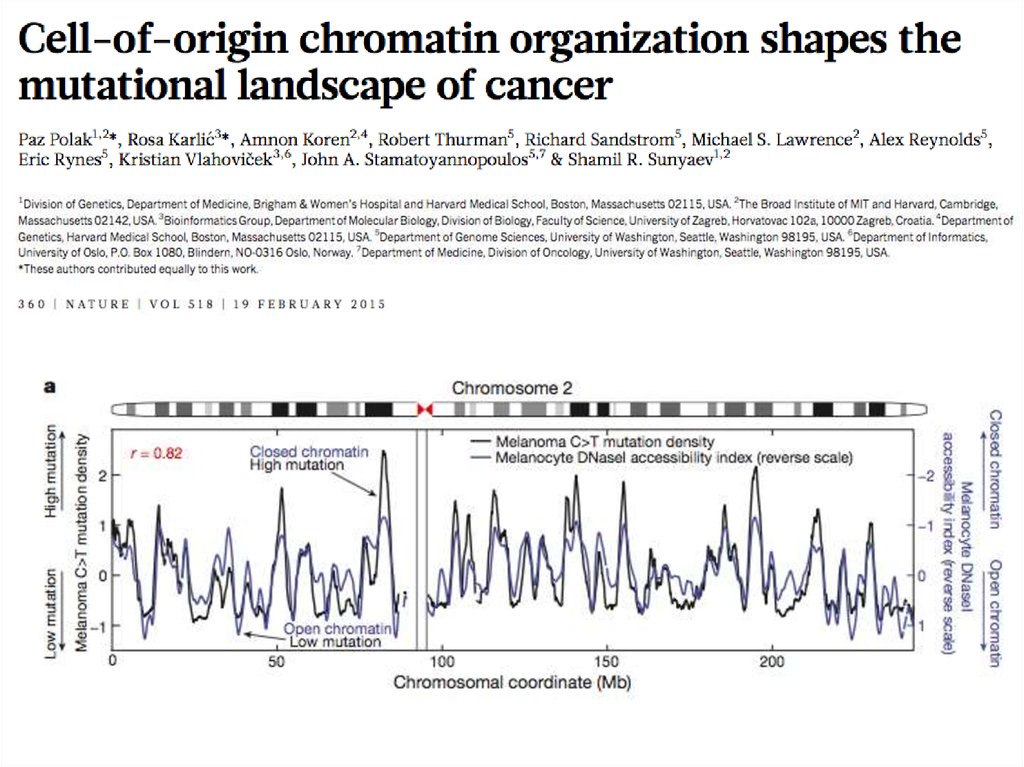
36.
Random Forest model predicts cancer mutation densities from epigenomicmark ups
37.
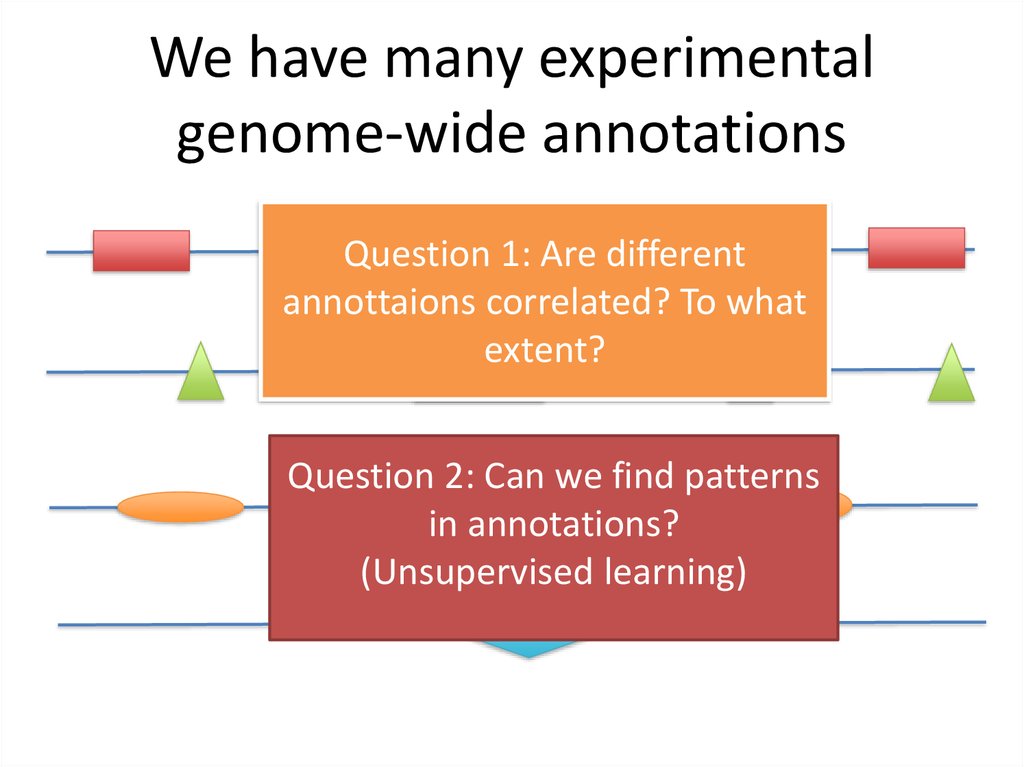
38. We have many experimental genome-wide annotations
Question 1: Are differentannottaions correlated? To what
extent?
Question 2: Can we find patterns
in annotations?
(Unsupervised learning)
39. Annotations under different conditions
40. Как много данных?
• Roadmap Epigenomics~ 3 000 полногеномных данных
• ENCODE Encyclopedia of Genomic Elements
~ 9000 полногеномных данных
• International Cancer Genome Consortium
~ 20 000 patients (~50 типов рака)
• The Cancer Genome Atlas
~ patients 11 000 (~33 типа рака)
41. Открытые вопросы
• Какие участки кода работают одновременно?• Как переключать режимы работы клетки?
• Как перепрограммируется код для разных
типов тканей?
• Сколько механизмов регуляции существует в
клетках (надежда на универсальность)?
















 Медицина
Медицина

