Похожие презентации:
Transcription in bacteria
1. Transcription in bacteria
MSU & SkolTechTranscription
in bacteria
1755
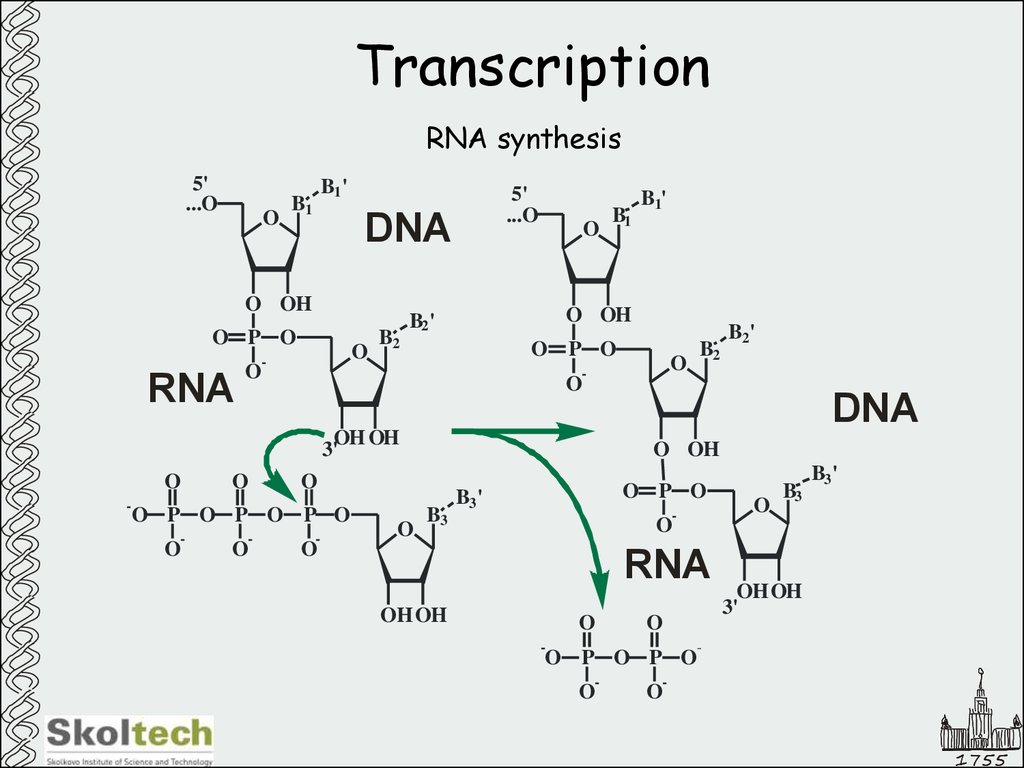
2. Transcription
RNA synthesis5'
...O
O
B1 '
B1
DNA
O OH
O P O
O
-
RNA
5'
...O
B2
O
O P O
O
-
O
O
-
O
B2
O
-
B2 '
O
DNA
O OH
O
O P O P O P O
-
B1'
O OH
B2 '
OH OH
3'
O
B1
-
O
B3
O P O
B3 '
-
O
B3
B3 '
O
O
RNA
OHOH
O
O
-
OH OH
3'
-
O P O P O
-
O
-
O
1755
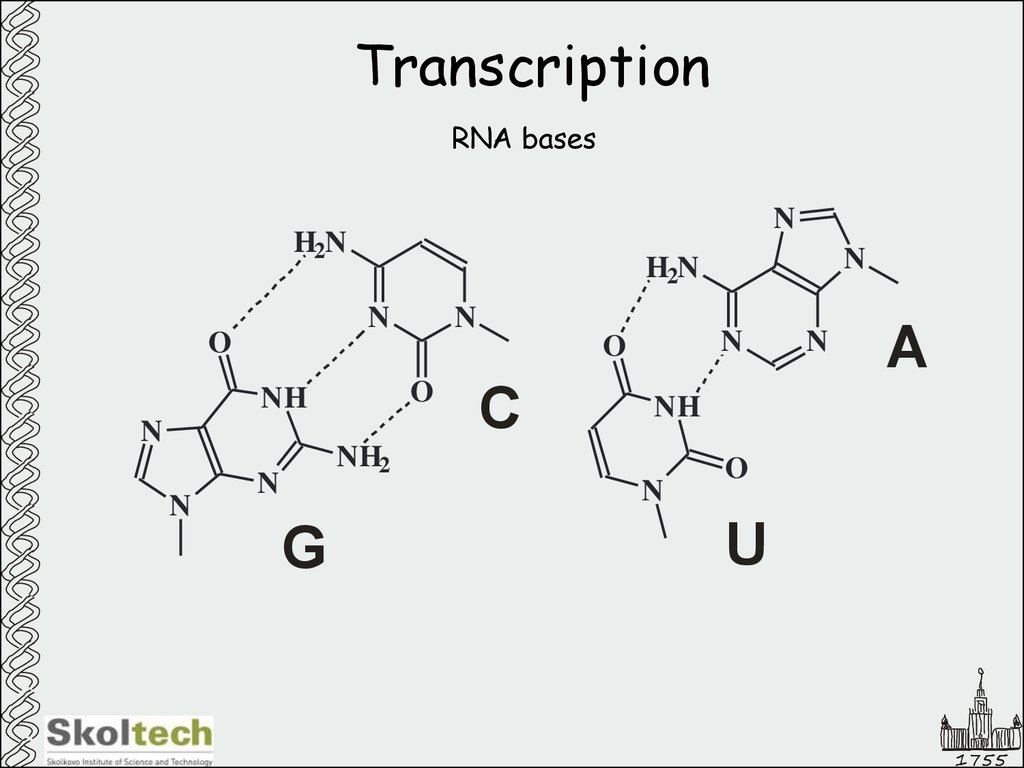
3. Transcription
RNA basesN
H2N
N
O
O
NH
N
N
N
O
N
C
N
G
N
A
NH
NH2
N
N
H2N
O
U
1755
4. Transcription
RNA polymerasea2bb’ws
Core
a 40 kD
b 155 kD
b’ 165 kD
w 10 kD
RNA polymerase
s 70 major
s 20 Fe2+ transport
s 28 flagella
s 32 heat shock
s 54 nitrogen limitation
s 24 periplasm stress
s 38 stationary phase
1755
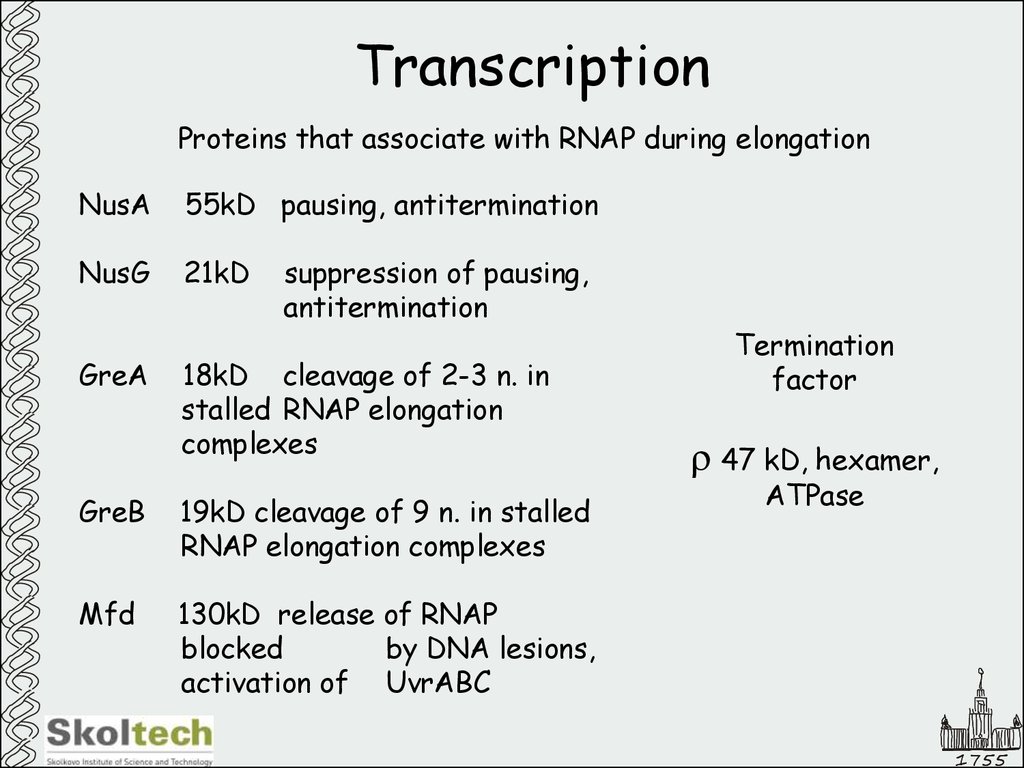
5. Transcription
Proteins that associate with RNAP during elongationNusA
55kD pausing, antitermination
NusG
21kD
GreA
suppression of pausing,
antitermination
18kD cleavage of 2-3 n. in
stalled RNAP elongation
complexes
GreB
19kD cleavage of 9 n. in stalled
RNAP elongation complexes
Mfd
130kD release of RNAP
blocked
by DNA lesions,
activation of UvrABC
Termination
factor
r 47 kD, hexamer,
ATPase
1755
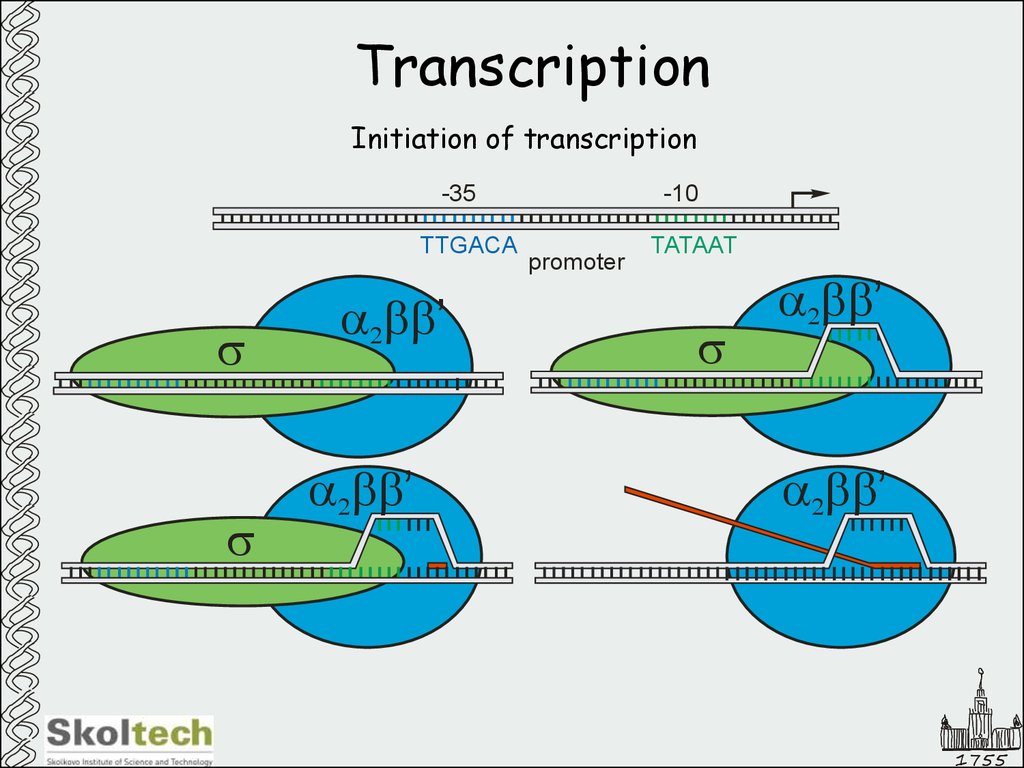
6. Transcription
Initiation of transcription-10
-35
TTGACA
s
s
a2bb’
a2bb’
promoter
TATAAT
s
a2bb’
a2bb’
1755
7. Transcription
Promoters and s factors-35
-10
E.coli s70
TTGACA
TATAAT
E.coli s32
TCTC-CCCTTGAA
CCCCAT-TA
E.coli s54
(-24)CTGG-A
(-12)TTGCA
B. sub sA
TTGACA
TATAAT
B. sub sB
AGGTTTAA
GGGTAT
B. sub sD
CTAAA
CCGATAT
B. sub sE
ATATT
ATACA
B. sub sK
AC
CATA---T
B. sub sH
CAGGA
GAATT—T
SPO1sgp28
AGGAGA
TTT-TTT
T4sgp55
-
TATAAATA
s
1755
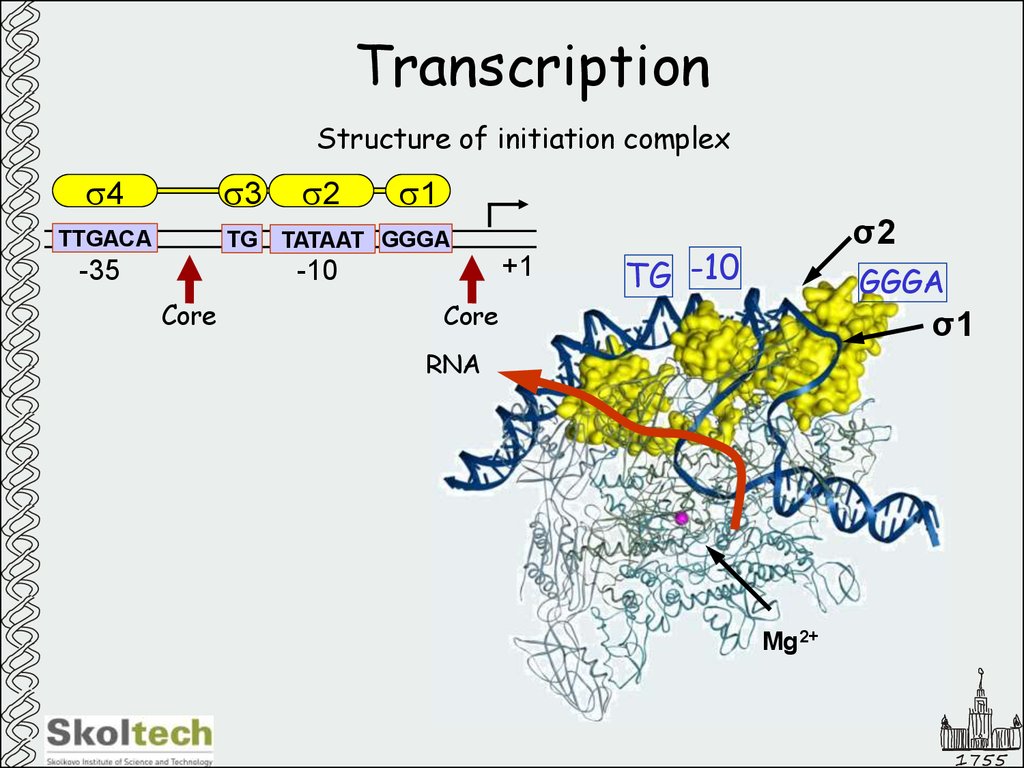
8. Transcription
Structure of initiation complexs4
s3
TTGACA
TG
-35
s2
s1
TATAAT GGGA
+1
-10
Core
Core
σ2
TG -10
GGGA
σ1
σ4
-35
Mg2+
1755
9. Transcription
Structure of initiation complexs4
s3
TTGACA
TG
-35
s2
s1
TATAAT GGGA
+1
-10
Core
Core
σ2
TG -10
GGGA
σ1
RNA
Mg2+
1755
10. Transcription
ElongationNusG
NusA
NusG
a2bb’
NusA
a2bb’
Pausing, regulation
or secondary structure
formation
Elongation
NusG
NusG
NusA
a2bb’
GreA/B
NusA
a2bb’
Mfd
TT
Release of stalled
complexes
Release of RNAP
stalled at DNA lesions
1755
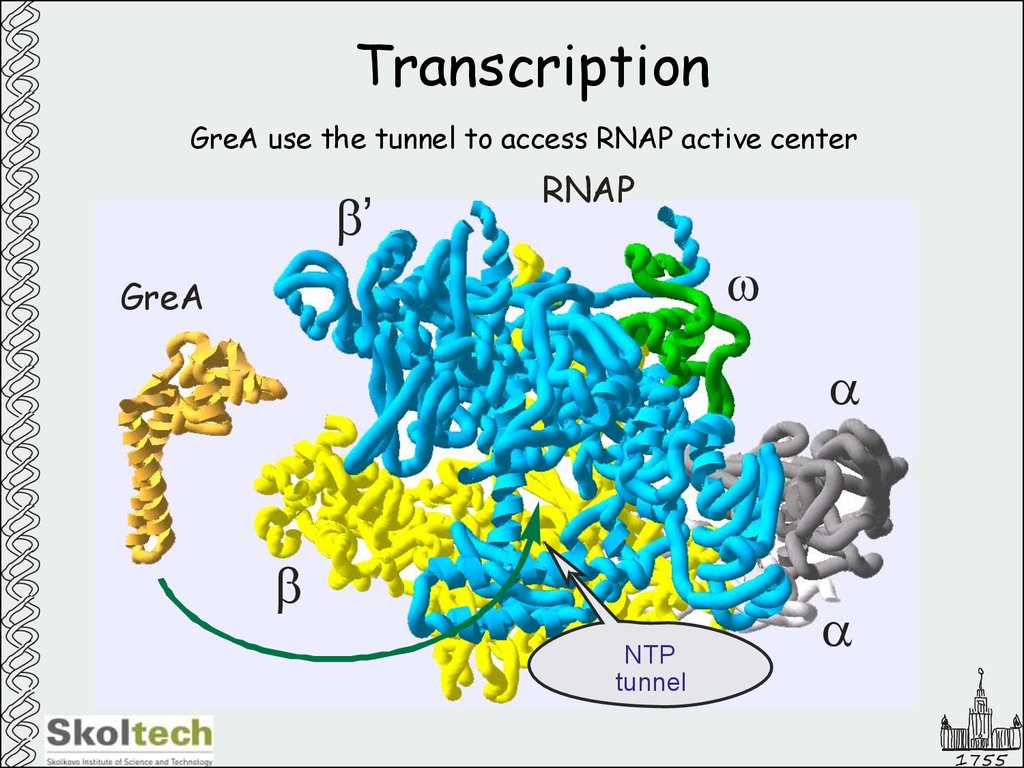
11. Transcription
GreA use the tunnel to access RNAP active centerb’
RNAP
w
GreA
a
b
NTP
tunnel
a
1755
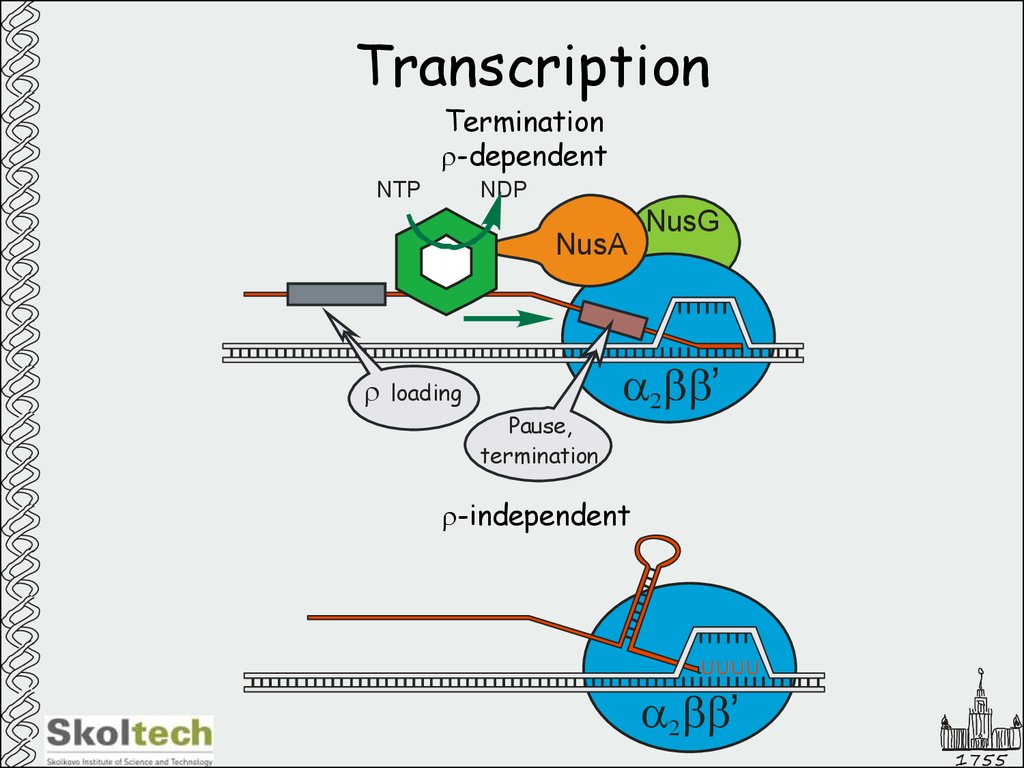
12. Transcription
Terminationr-dependent
NTP
NDP
NusA
r
loading
Pause,
termination
NusG
a2bb’
r-independent
UUUU
a2bb’
1755
13. Transcription
r-factor1755
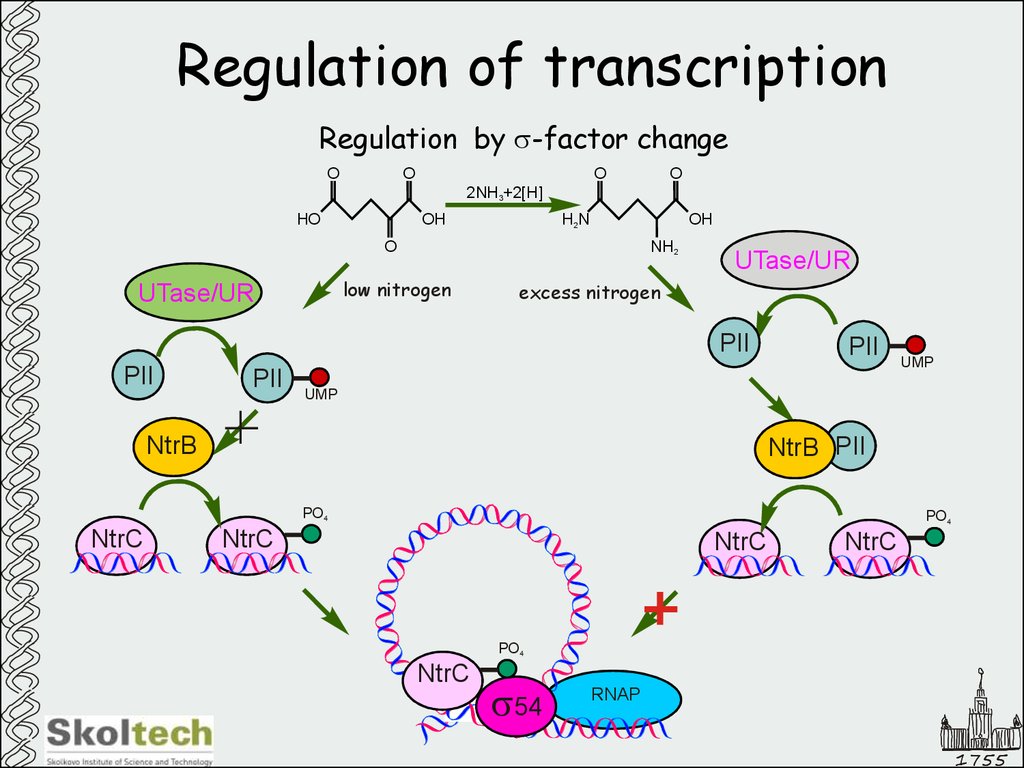
14. Regulation of transcription
Regulation by s-factor changeO
O
O
O
2NH3+2[H]
HO
OH
H2N
OH
O
NH2
low nitrogen
UTase/UR
UTase/UR
excess nitrogen
PII
PII
PII
PII
UMP
NtrB
NtrB PII
PO4
NtrC
UMP
PO4
NtrC
NtrC
NtrC
PO4
NtrC
s54
RNAP
1755
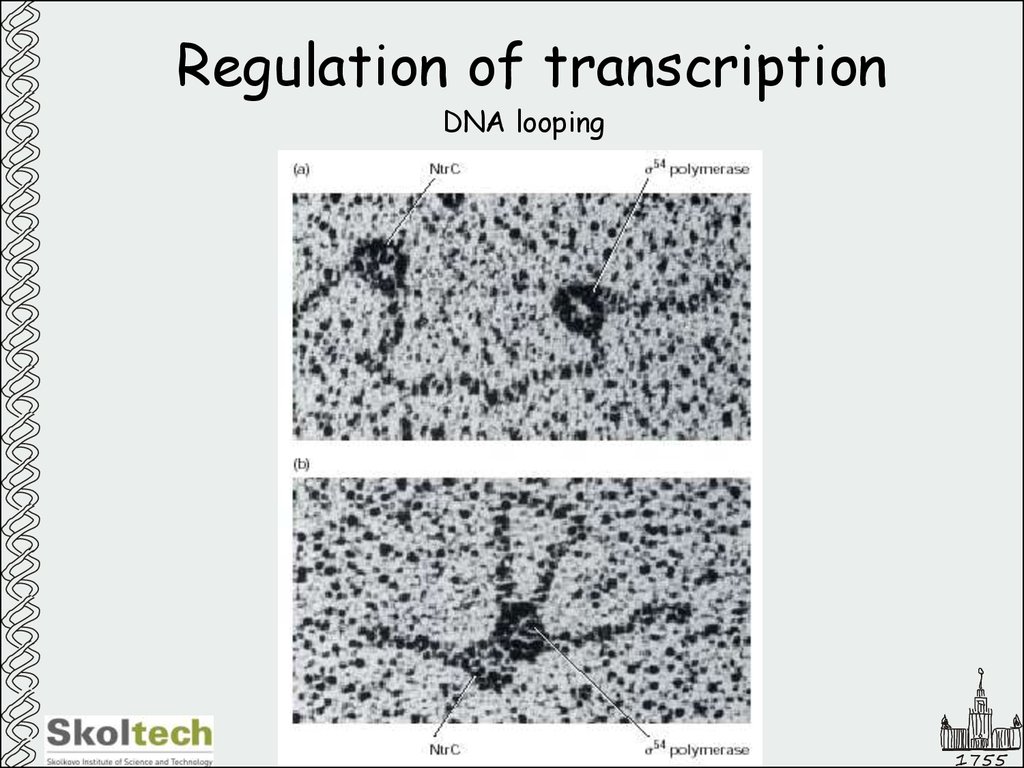
15. Regulation of transcription
DNA looping1755
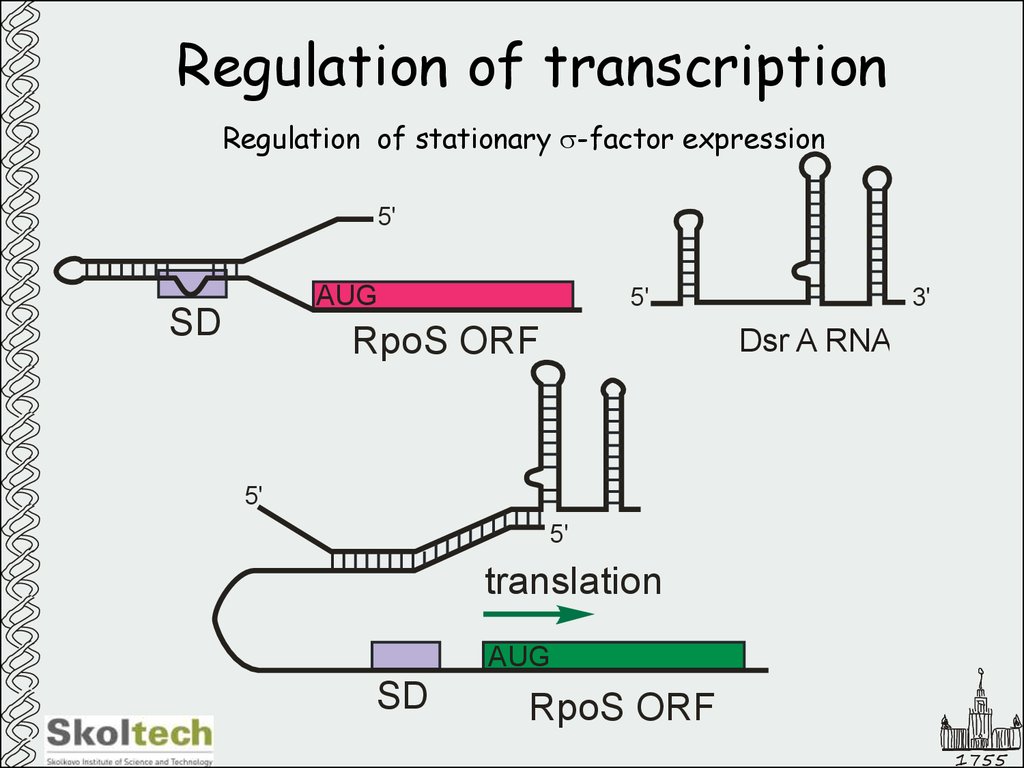
16. Regulation of transcription
Regulation of stationary s-factor expression5'
AUG
SD
5'
RpoS ORF
3'
Dsr A RNA
5'
5'
translation
AUG
SD
RpoS ORF
1755
17. Regulation of transcription
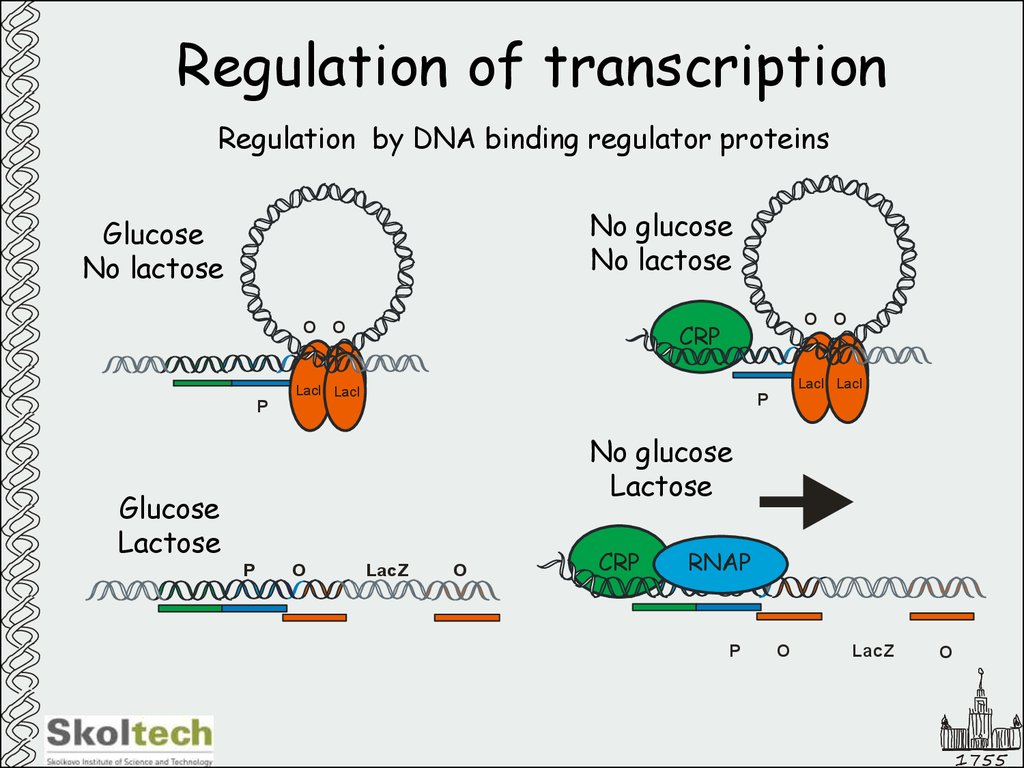
Regulation by DNA binding regulator proteinslac operon:
Glucose is preferable
energy source
Lactose could be utilized
if available
Jacob, Monod
Nobel Prize
1755
18. Regulation of transcription
Regulation by DNA binding regulator proteinsNo glucose
No lactose
Glucose
No lactose
O O
CRP
O O
LacI LacI
LacI LacI
P
P
No glucose
Lactose
Glucose
Lactose
P
O
LacZ
O
CRP
RNAP
P
O
LacZ
O
1755
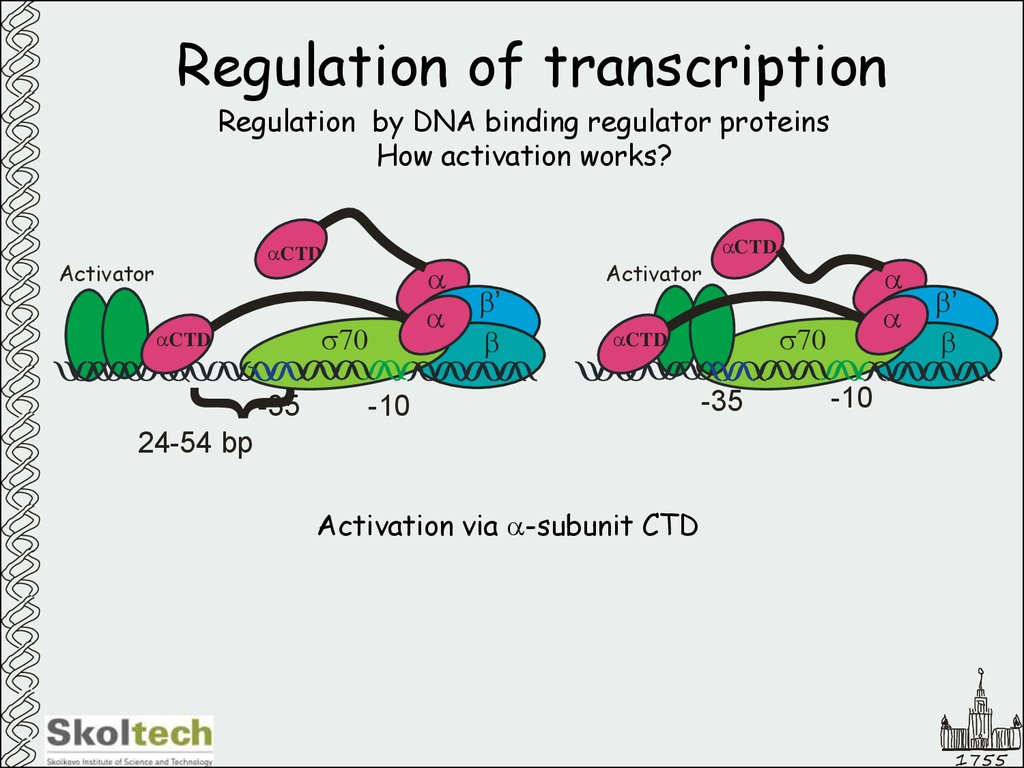
19. Regulation of transcription
Regulation by DNA binding regulator proteinsHow activation works?
aCTD
Activator
s70
aCTD
}
-35
a
a
b’
b
Activator
aCTD
s70
aCTD
-10
a
a
-35
b’
b
-10
24-54 bp
Activation via a-subunit CTD
1755
20. Regulation of transcription
Regulation by DNA binding regulator proteinsHow activation works?
s70
a
a
-10
-35
MerR
b’
b
s70
-35 2+
Hg
a
a
b’
b
-10
2+
Hg
MerR
Activation/repression via DNA structure change
1755
21. Regulation of transcription
Regulation by DNA binding regulator proteinsComplex regulation
Nir-operon
Fis
No oxygen
IHF
FNR FNR
-35
Nitrite ions present
-10
No oxygen
IHF
aCTD
Fis
NarL/NarP
FNR
-35
a
a
s70
b’
b
-10
1755
22. Regulation of transcription
Regulation by DNA binding regulator proteinsTranscription factor activity change by localization
MBP
MalG
MBP
MalF
MalK MalK
ATP
ATP
MalT
MalG
MalK
ADP
MalT
MalF
MalK
ADP
transcription
1755
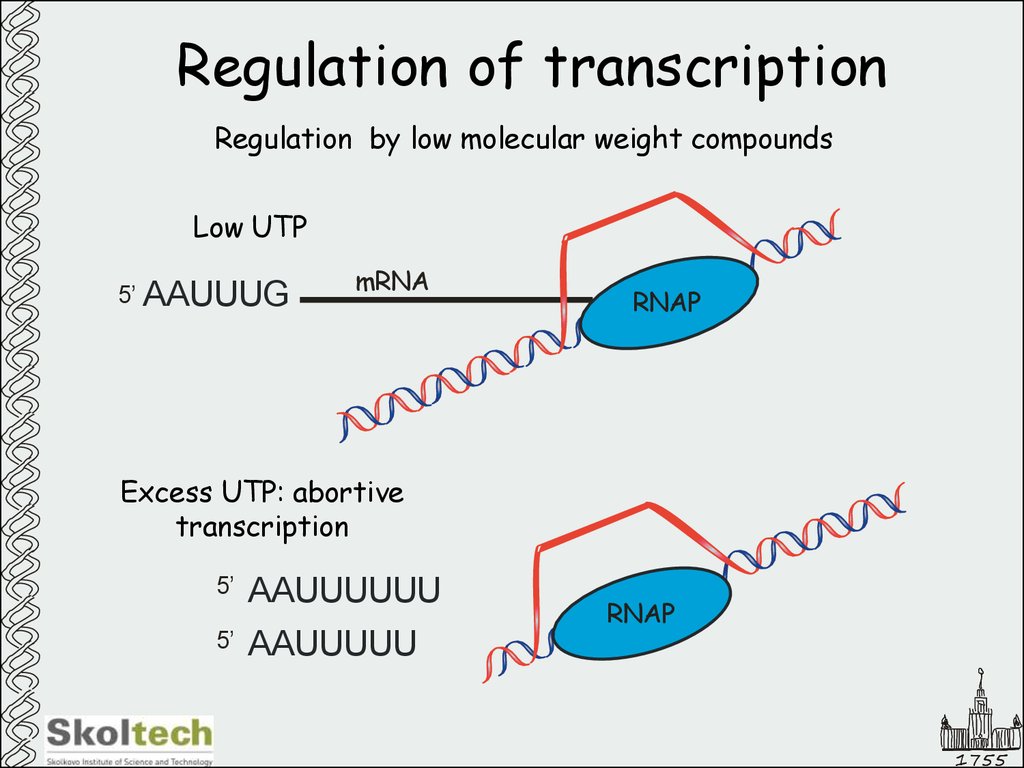
23. Regulation of transcription
Regulation by low molecular weight compoundsLow UTP
5’ AAUUUG
mRNA
RNAP
Excess UTP: abortive
transcription
5’
AAUUUUUU
5’
AAUUUUU
RNAP
1755
24. Regulation of transcription
Regulation by low molecular weight compoundsppGpp
DksA
GreA
O
N
O
-
O
O
P O P O
-
-
O
N
O
O
-
O
-
O
O
NH
NH 2
OH
-
P O P O
O
O
1755
25. Regulation of transcription
Regulation of elongation and terminationAttenuation of E. coli Trp operon
Low Trp
concentration
ribosome
mRNA
5’
1
2
3
a2bb’
4
UGG UGG
High Trp
concentration
ribosome
3
mRNA
5’
1
UGG UGG
2
4
UUUUUUUUUUU
1755
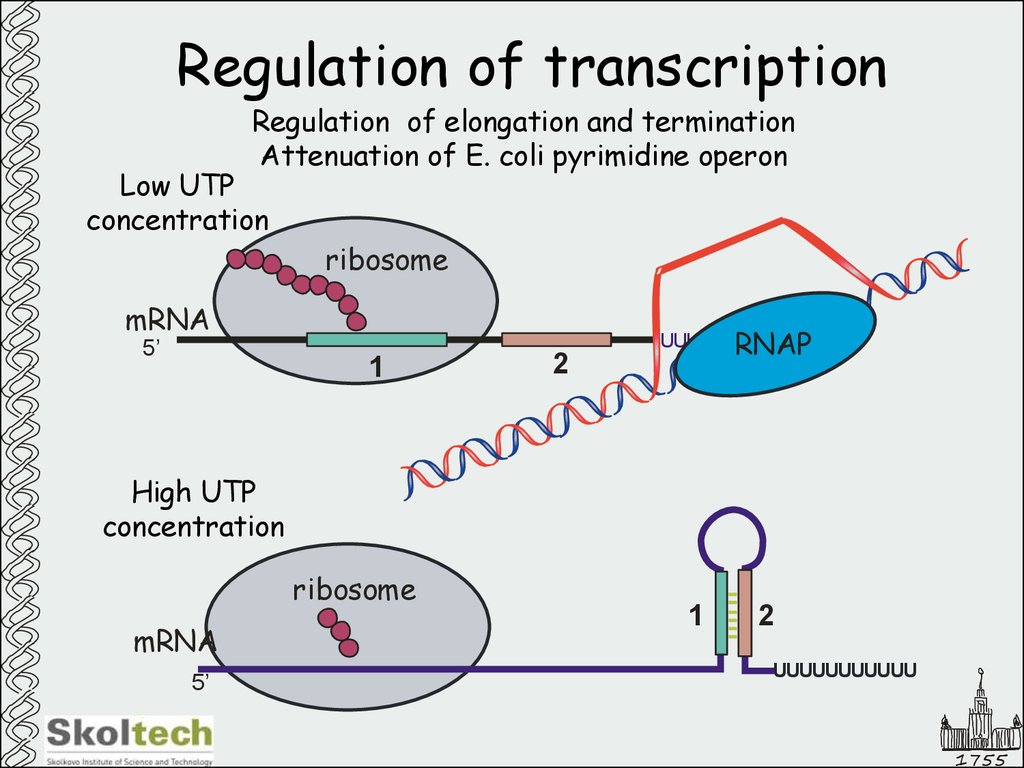
26. Regulation of transcription
Regulation of elongation and terminationAttenuation of E. coli pyrimidine operon
Low UTP
concentration
ribosome
mRNA
5’
UUUUU
1
2
RNAP
High UTP
concentration
ribosome
mRNA
5’
1
2
UUUUUUUUUUU
1755
27. Regulation of transcription
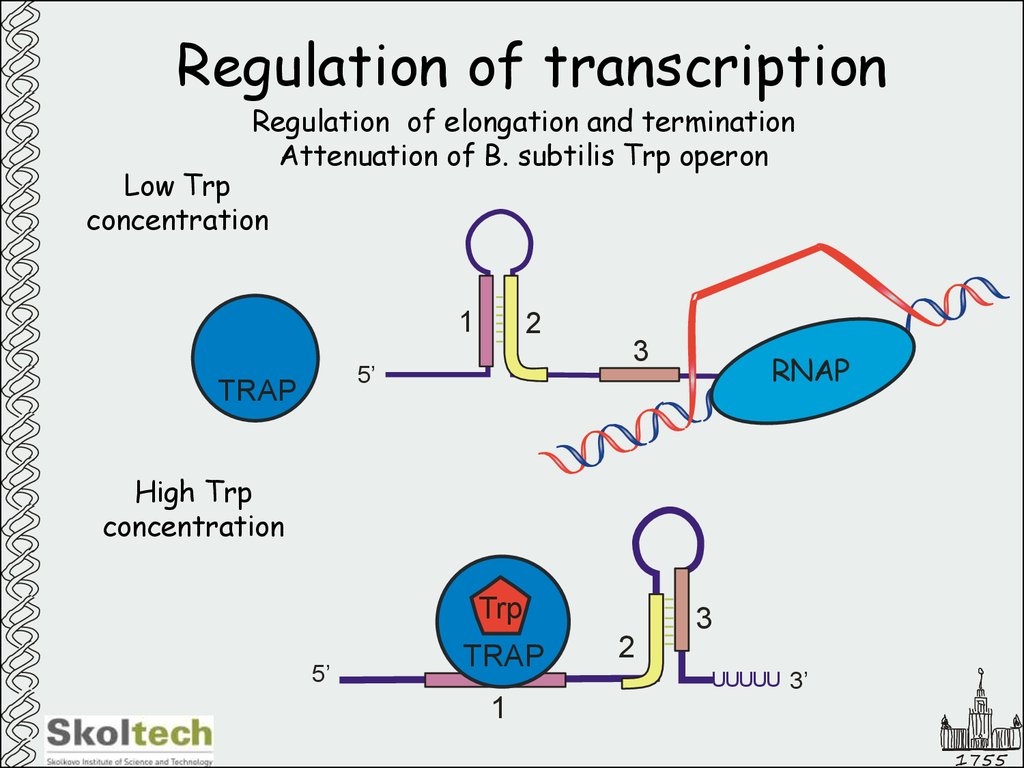
Regulation of elongation and terminationAttenuation of B. subtilis Trp operon
Low Trp
concentration
1
2
3
5’
TRAP
RNAP
High Trp
concentration
Trp
5’
TRAP
3
2
UUUUU 3’
1
1755
28. Regulation of transcription
Regulation of elongation and terminationAttenuation of B. subtilis TyrRS gene
transcription
termination
of Tyr-ARS
UUUUUU
Tyr-тРНК
Tyr
High
aminoacylation
тРНК
Tyr
Low
aminoacylation
1755
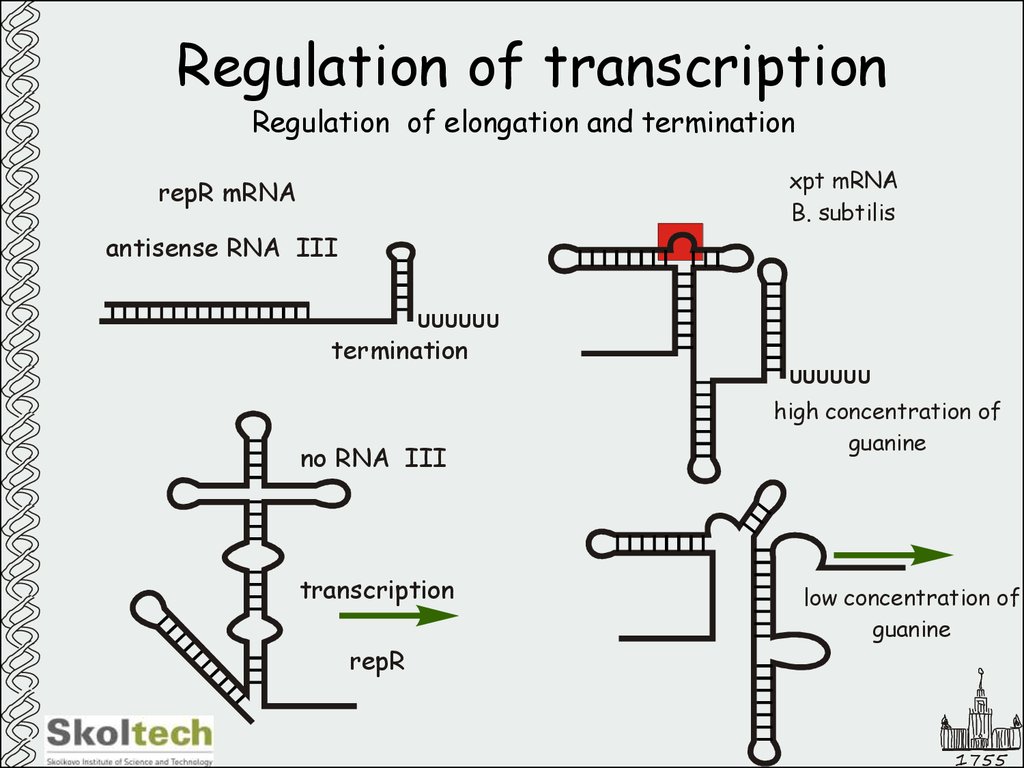
29. Regulation of transcription
Regulation of elongation and terminationxpt mRNA
B. subtilis
repR mRNA
antisense RNA III
UUUUUU
termination
UUUUUU
no RNA III
transcription
repR
high concentration of
guanine
low concentration of
guanine
1755
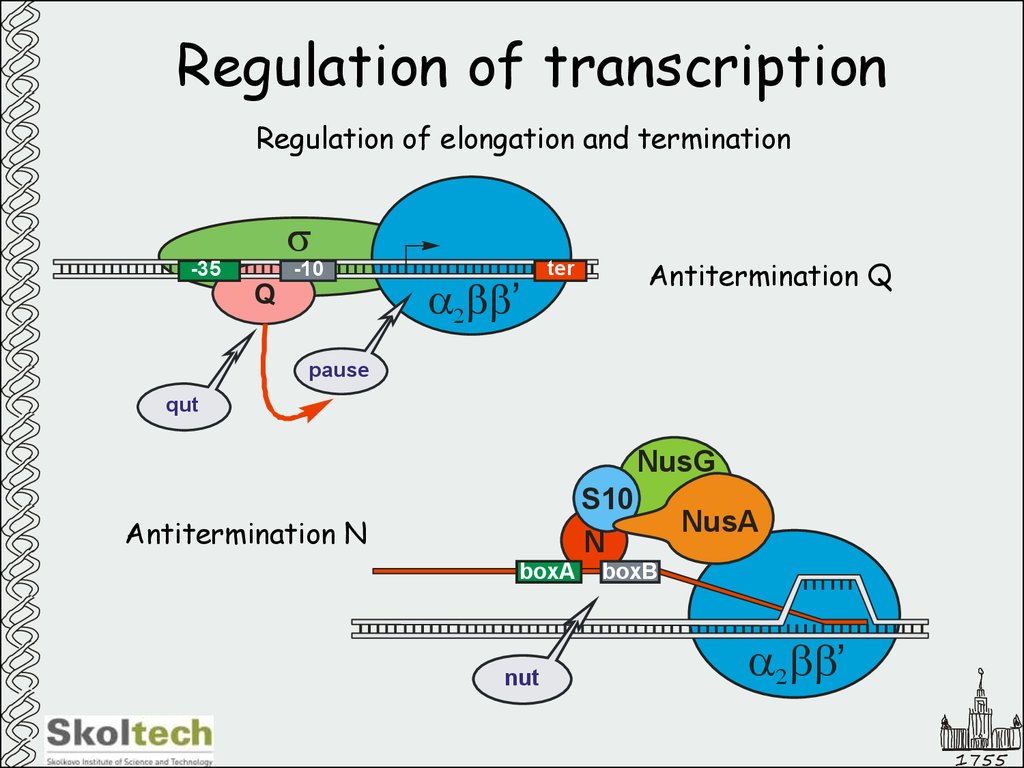
30. Regulation of transcription
Regulation of elongation and terminations
-35
-10
Q
a2bb’
Antitermination Q
ter
pause
qut
NusG
S10
N
Antitermination N
boxA
nut
NusA
boxB
a2bb’
1755
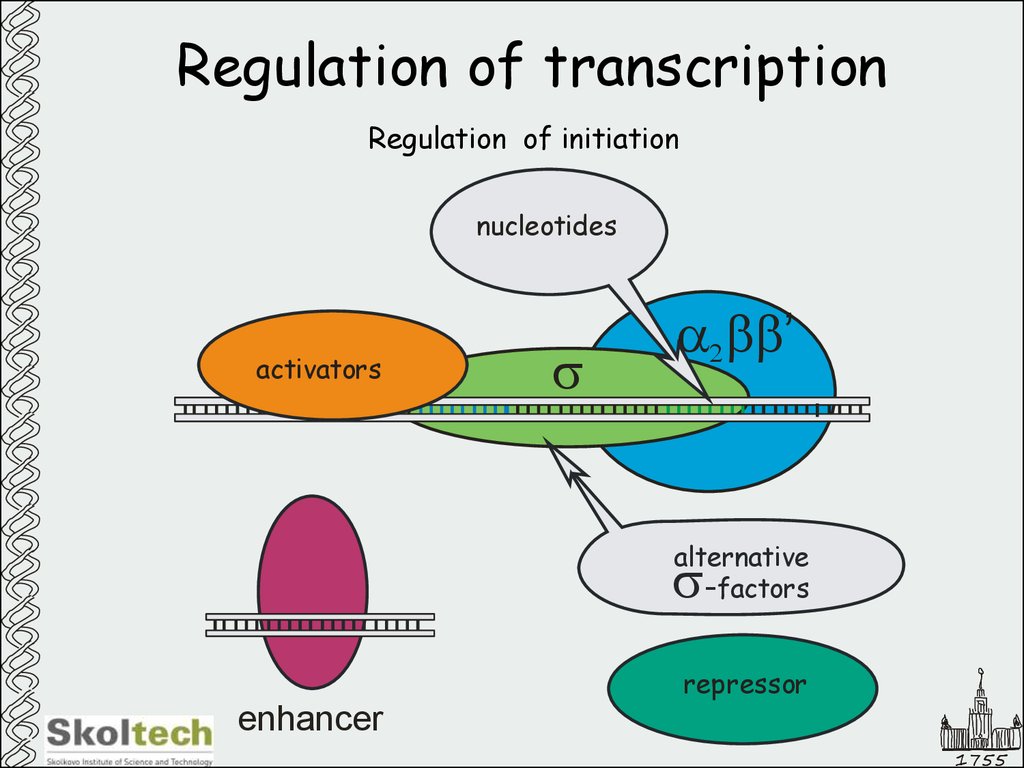
31. Regulation of transcription
Regulation of initiationnucleotides
activators
s
a2bb’
alternative
-factors
s
repressor
enhancer
1755
32. Regulation of transcription
Regulation of elongation and terminationAttenuation
1
2
3
N
NusG
a
bb
’
2
NusA
a2bb’
- translation speed
-transcription speed
-protein binding
-RNA binduing
-small molecule binding
Antitermination
- RNA binding protein
antiterminator
- DNA binding protein
antiterminator
1755
33. Examples of the transcriptional control
Bacterial flagella biosynthesisFlagella is expensive and energy consuming machinery
1755
34. Examples of the transcriptional control
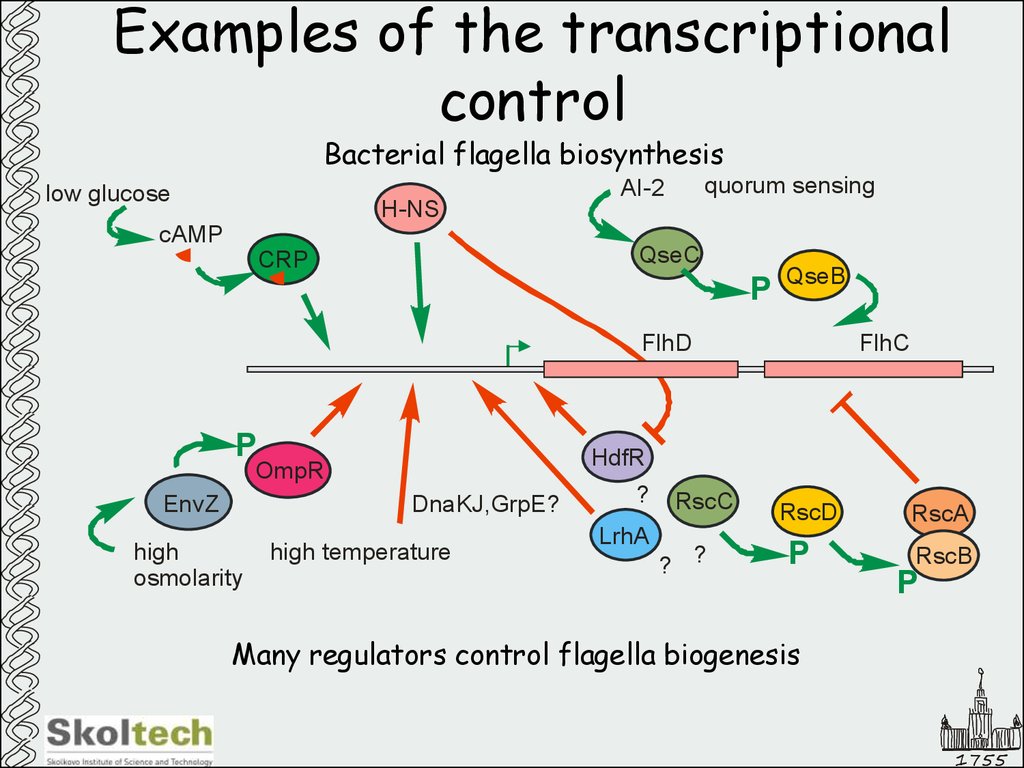
Bacterial flagella biosynthesislow glucose
H-NS
cAMP
quorum sensing
AI-2
QseC
CRP
P
QseB
FlhD
P
HdfR
OmpR
EnvZ
DnaKJ,GrpE?
high
osmolarity
FlhC
high temperature
?
LrhA
RscC
? ?
RscD
P
RscA
RscB
P
Many regulators control flagella biogenesis
1755
35. Examples of the transcriptional control
Bacterial flagella biosynthesisFlhD4C2
s28
s70
s28
FlgM
FliA(s28)
FlhD4C2
FlgM
s
70
FlgM
hook
basal body
FlhDC activates a set of early promoters
1755
36. Examples of the transcriptional control
Bacterial flagella biosynthesisFlgM
FlhD4C2
s
FlgM
28
hook
basal body
FlgM
FlhD4C2
s28
s28
FliA(s )
28
s
28
FliС(flagellin)
Positive feedback loop starts after hook/basal body assembly
1755
37. Examples of the transcriptional control
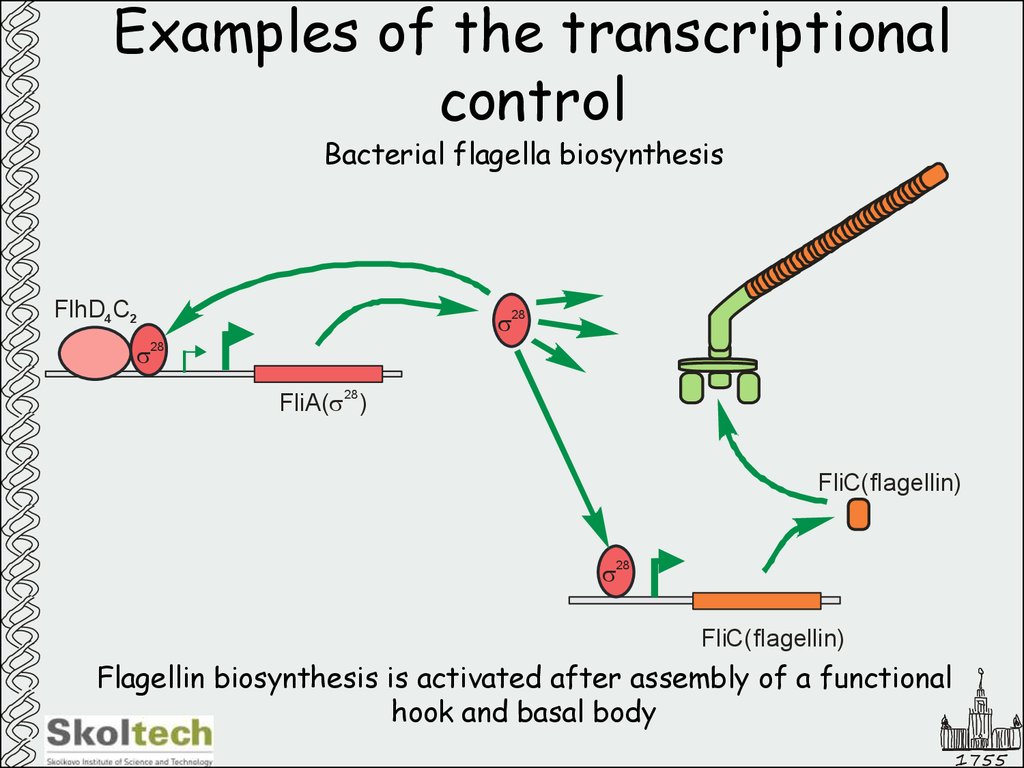
Bacterial flagella biosynthesisFlhD4 C2
s
28
s28
FliA(s )
28
FliС( flagellin)
s28
FliС( flagellin)
Flagellin biosynthesis is activated after assembly of a functional
hook and basal body
1755
38. Examples of the transcriptional control
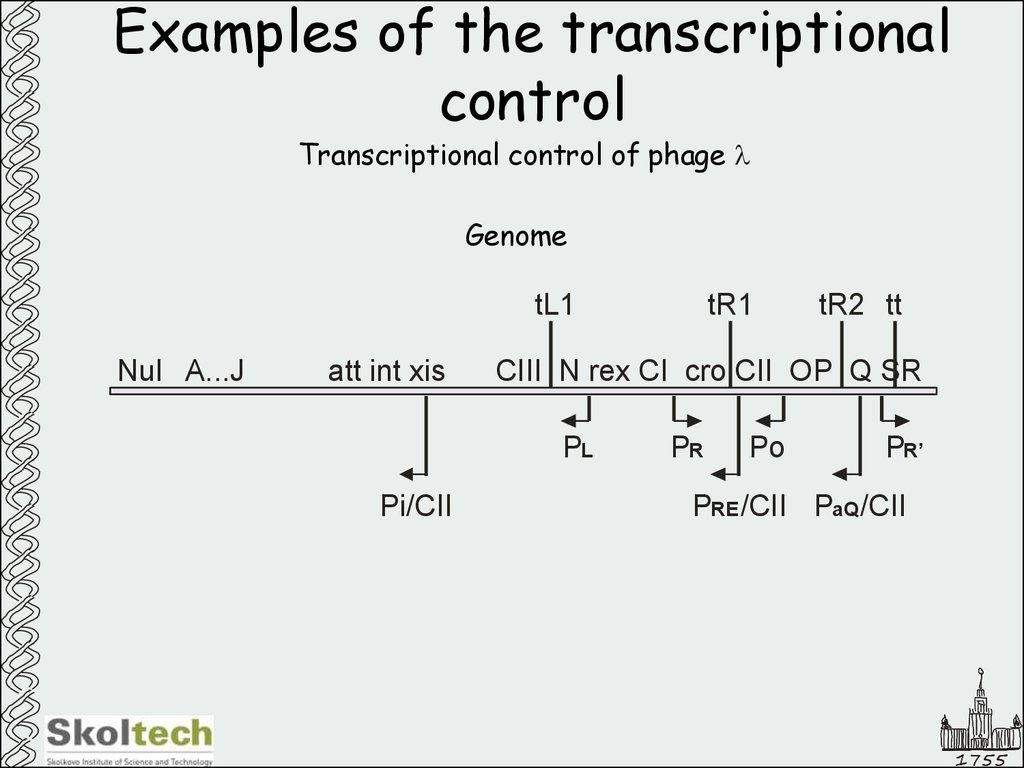
Transcriptional control of phage lGenome
tL1
NuI A...J
att int xis
tR2 tt
CIII N rex CI cro CII OP Q SR
PL
Pi/CII
tR1
PR
Po
PR’
PRE /CII PaQ /CII
1755
39. Examples of the transcriptional control
Transcriptional control of phage lEarly genes
tL1
NuI A...J
att int xis
tR1
tR2 tt
CIII N rex CI cro CII OP Q SR
PL
Pi/CII
PR
Po
PR’
PRE /CII PaQ /CII
Making a choice
Hunger
cAMP
HflA
CII
Lysogeny
1755
40. Examples of the transcriptional control
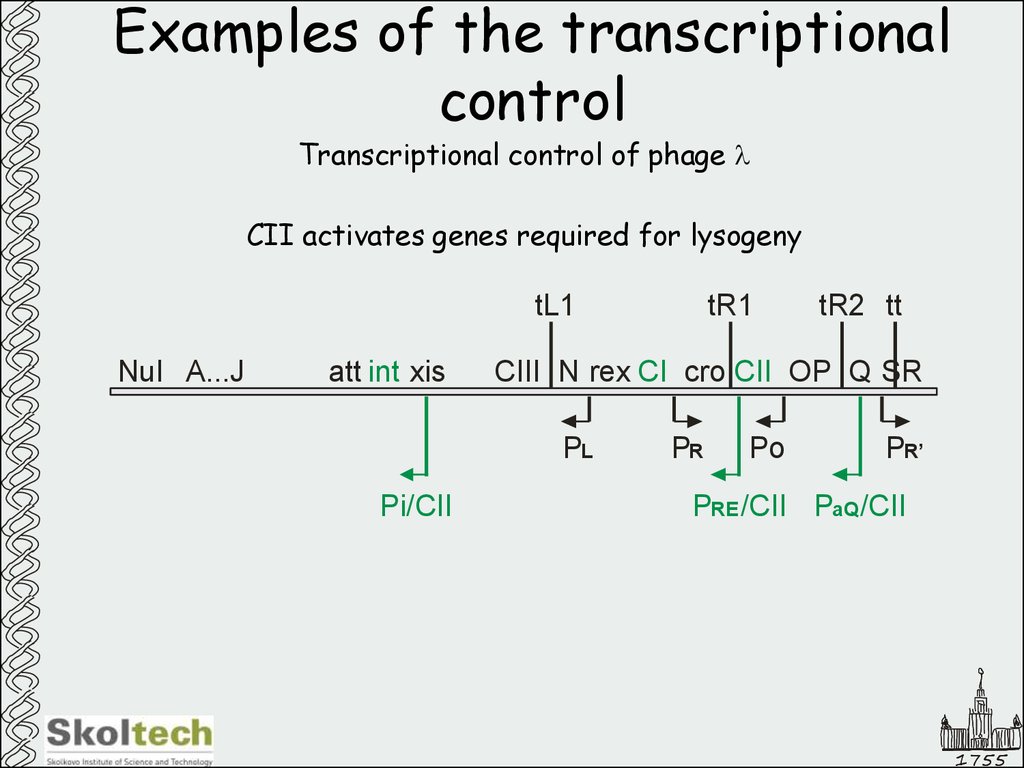
Transcriptional control of phage lCII activates genes required for lysogeny
tL1
NuI A...J
att int xis
tR2 tt
CIII N rex CI cro CII OP Q SR
PL
Pi/CII
tR1
PR
Po
PR’
PRE /CII PaQ /CII
1755
41. Examples of the transcriptional control
Transcriptional control of phage lCI inactivate everything but its own gene
tL1
NuI A...J
att int xis
tR1
CIII N rex CI cro CII OP Q SR
PL PRM PR
Pi/CII
tR2 tt
Po
PR’
PRE /CII PaQ /CII
1755
42. Examples of the transcriptional control
Transcriptional control of phage lCI protein: positive and negative feedback loops
PRM
RNAP
CI
ORIII ORII
PRM
CI
CI
ORIII ORII
PR
CI
ORI
PR
CI
ORI
1755
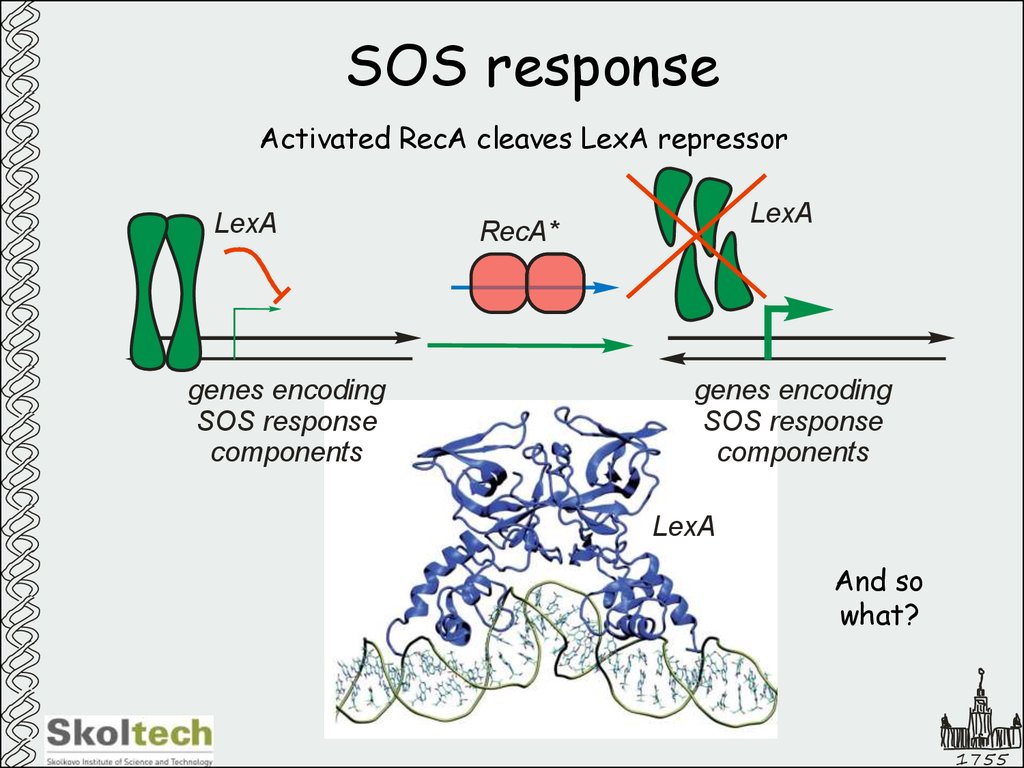
43. SOS response
Activated RecA cleaves LexA repressorLexA
genes encoding
SOS response
components
LexA
RecA*
genes encoding
SOS response
components
LexA
And so
what?
1755
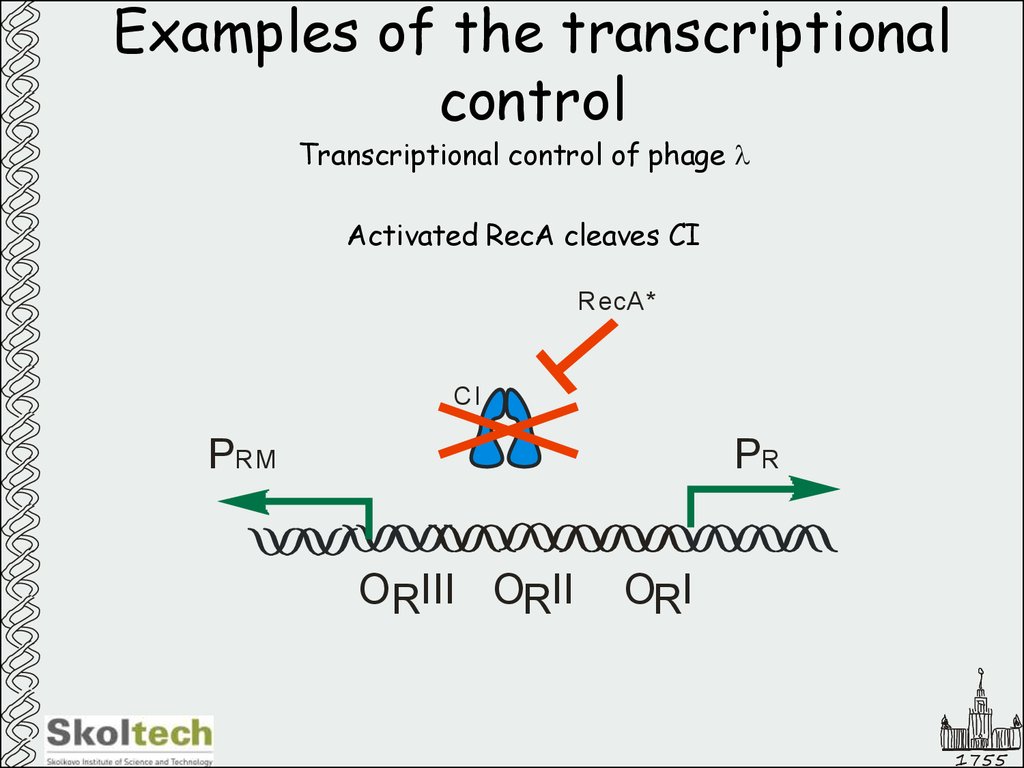
44. Examples of the transcriptional control
Transcriptional control of phage lActivated RecA cleaves CI
RecA*
CI
PRM
PR
ORIII ORII
ORI
1755
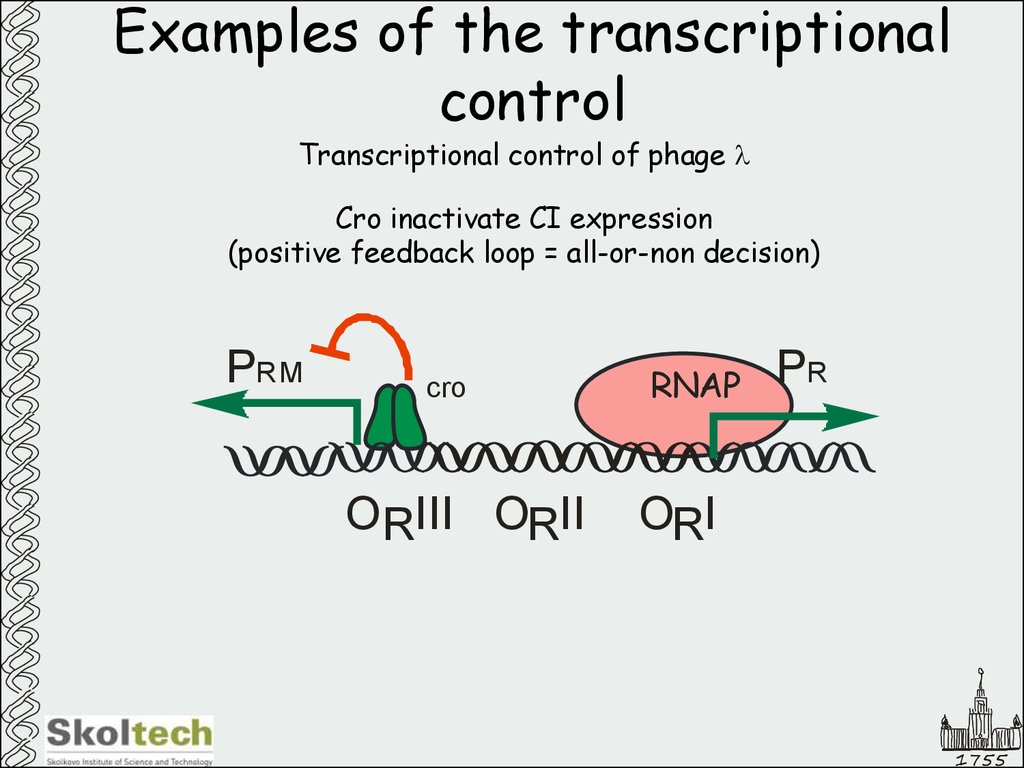
45. Examples of the transcriptional control
Transcriptional control of phage lCro inactivate CI expression
(positive feedback loop = all-or-non decision)
PRM
cro
ORIII ORII
RNAP
PR
ORI
1755
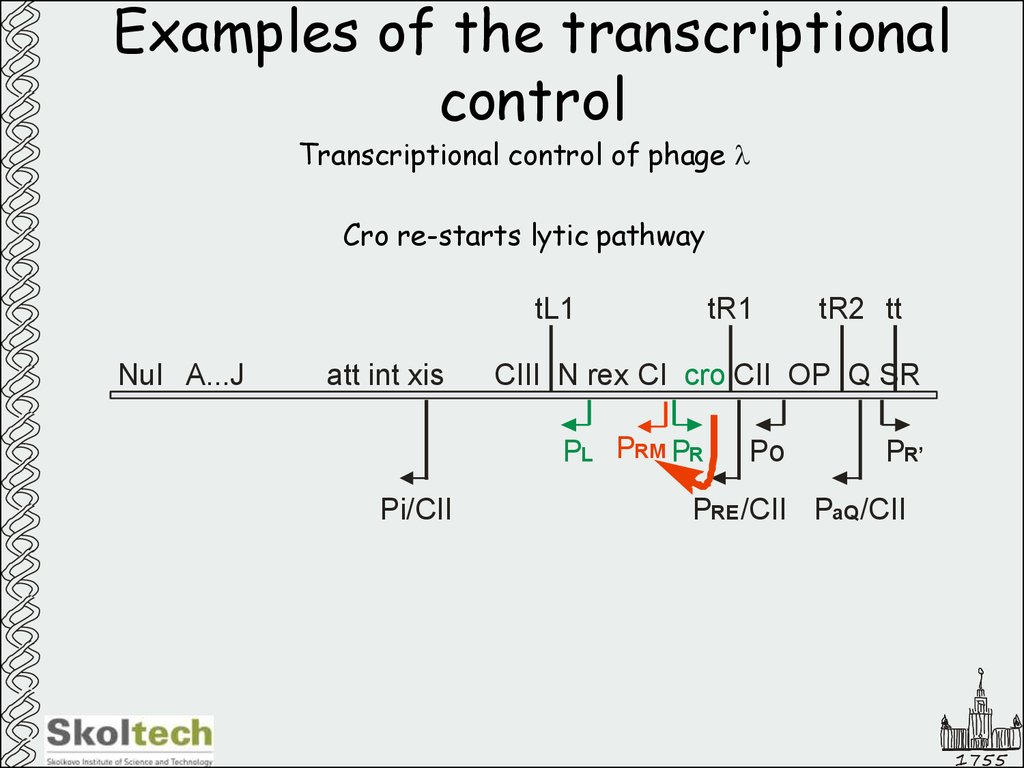
46. Examples of the transcriptional control
Transcriptional control of phage lCro re-starts lytic pathway
tL1
NuI A...J
att int xis
tR1
CIII N rex CI cro CII OP Q SR
PL PRM PR
Pi/CII
tR2 tt
Po
PR’
PRE /CII PaQ /CII
1755
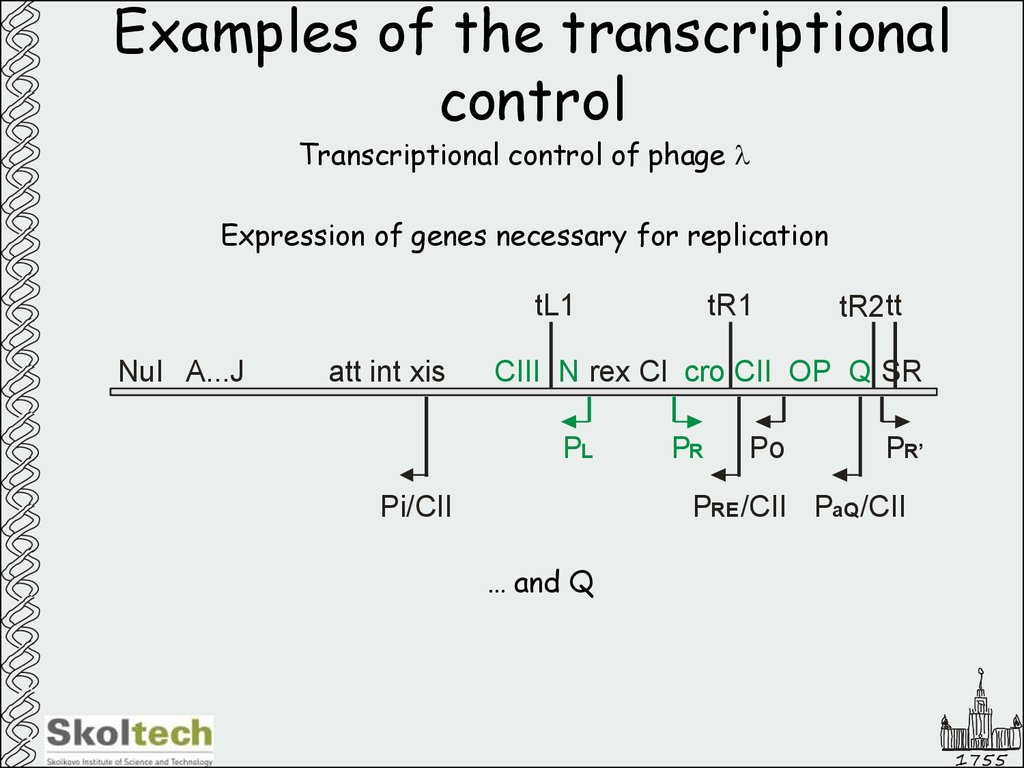
47. Examples of the transcriptional control
Transcriptional control of phage lExpression of genes necessary for replication
tL1
NuI A...J
att int xis
tR1
tR2tt
CIII N rex CI cro CII OP Q SR
PL
Pi/CII
PR
Po
PR’
PRE /CII PaQ /CII
… and Q
1755
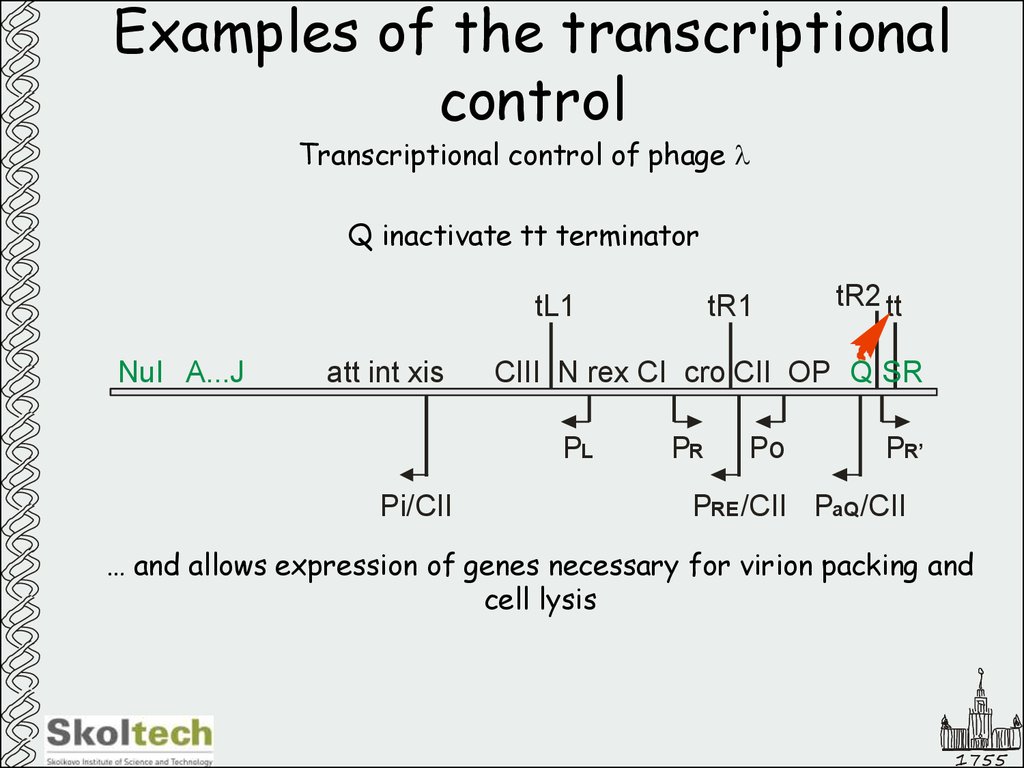
48. Examples of the transcriptional control
Transcriptional control of phage lQ inactivate tt terminator
tL1
NuI A...J
att int xis
tR2 tt
CIII N rex CI cro CII OP Q SR
PL
Pi/CII
tR1
PR
Po
PR’
PRE /CII PaQ /CII
… and allows expression of genes necessary for virion packing and
cell lysis
1755
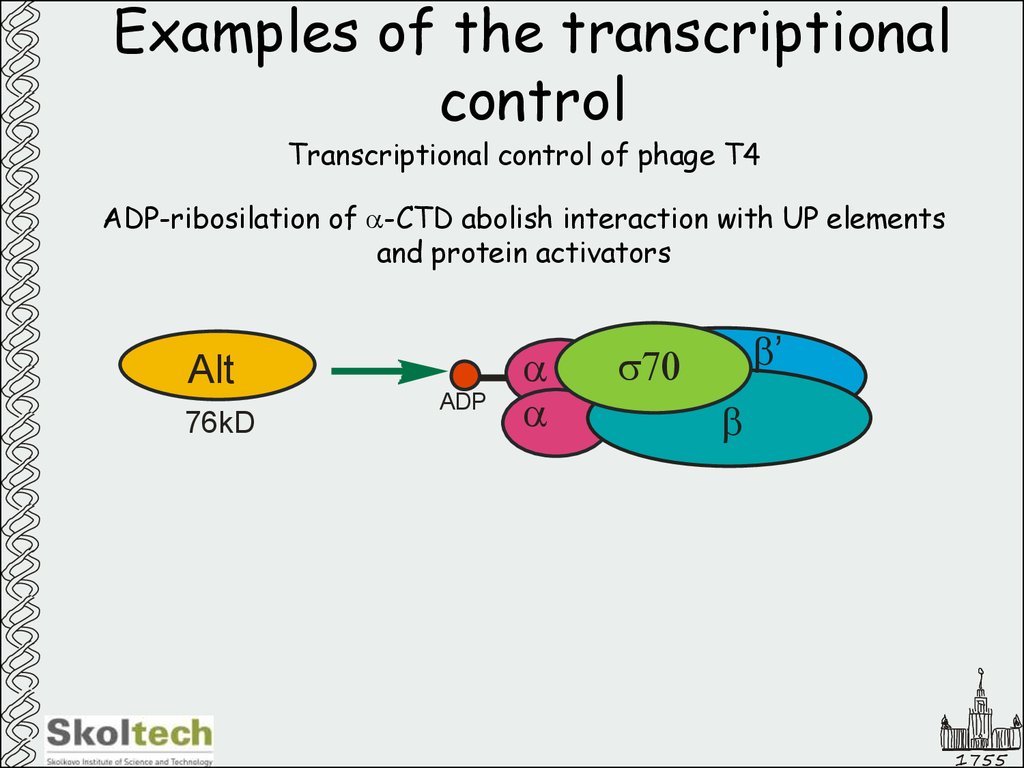
49. Examples of the transcriptional control
Transcriptional control of phage T4ADP-ribosilation of a-CTD abolish interaction with UP elements
and protein activators
Alt
76kD
ADP
a
a
b’
s70
b
1755
50. Examples of the transcriptional control
Transcriptional control of phage T4-Further ADP-ribosylation of a-CTD
- AsiA inhibit -35 box recognition
-MotA interacts with middle gene promoters
Mod
-Alc blocks transcription on DNA without 5hmC
AsiA
s70
MotA
mot
box
TATA
24kD
a
a
b’
b
ADP
ADP
Alc
19kD
1755
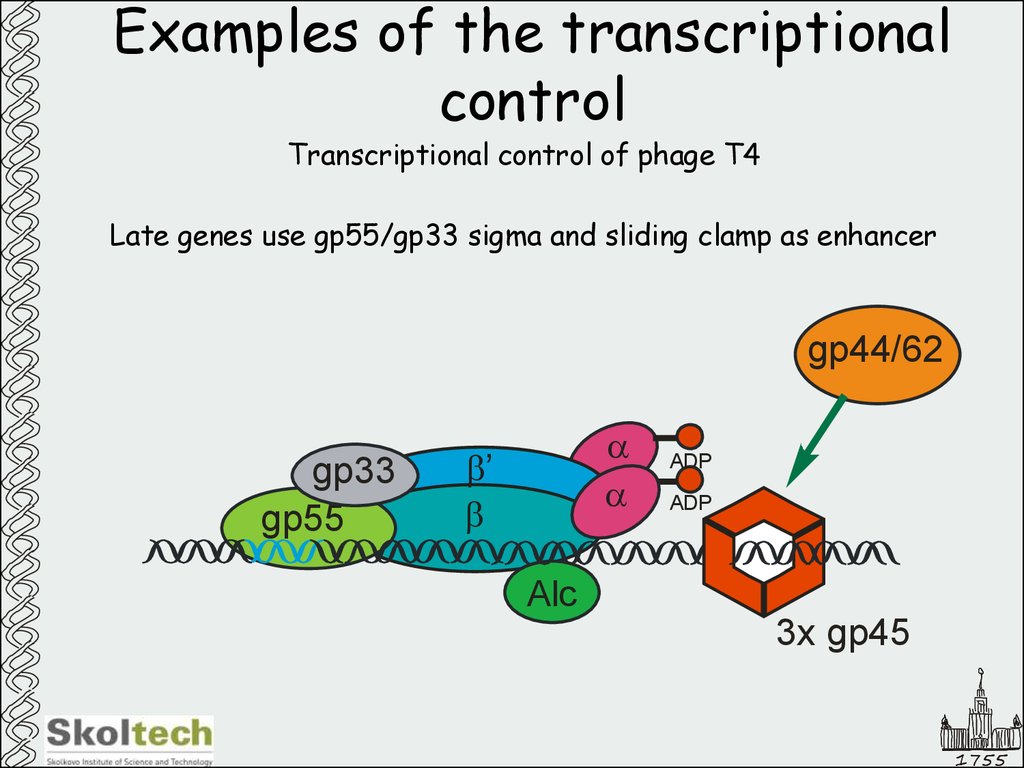
51. Examples of the transcriptional control
Transcriptional control of phage T4Late genes use gp55/gp33 sigma and sliding clamp as enhancer
gp44/62
gp33
gp55
a
a
b’
b
ADP
ADP
Alc
3x gp45
1755
52. Examples of the transcriptional control
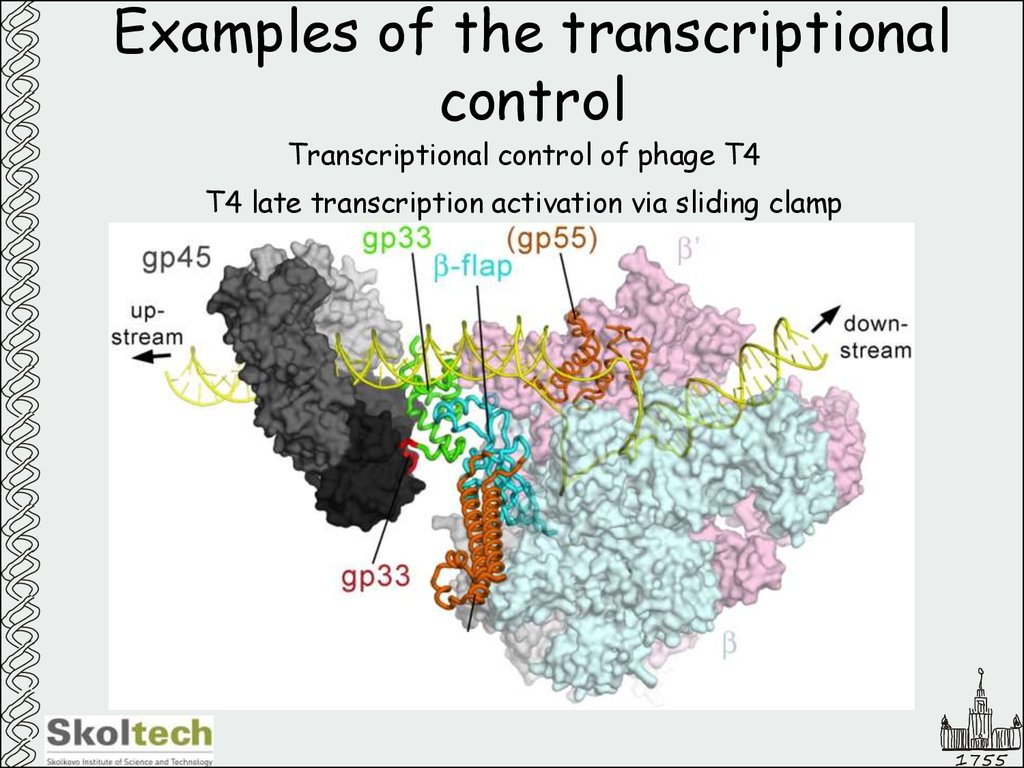
Transcriptional control of phage T4T4 late transcription activation via sliding clamp
1755



















 Химия
Химия








