Похожие презентации:
Twinscan-mars
1. Twinscan - MARS
Paul FlicekWashington University
EBI
2. Twinscan
Twinscan incorporates a pair wise alignmentnet (with an informant sequence) into its
algorithm
Twinscan “works” for human gene prediction
with almost any mammalian informant (but
mice and rats are the most useful)
Prediction characteristics are dependent on
the informant
3. Goals & Methods
Goals & MethodsExploit the differences in gene characteristics
resulting from each individual informant
genome
Create closely-related prediction by
incorporating mixtures of pair-wise alignment
nets into Twinscan
Cluster these predictions as a way to find
multiple transcripts from a single gene
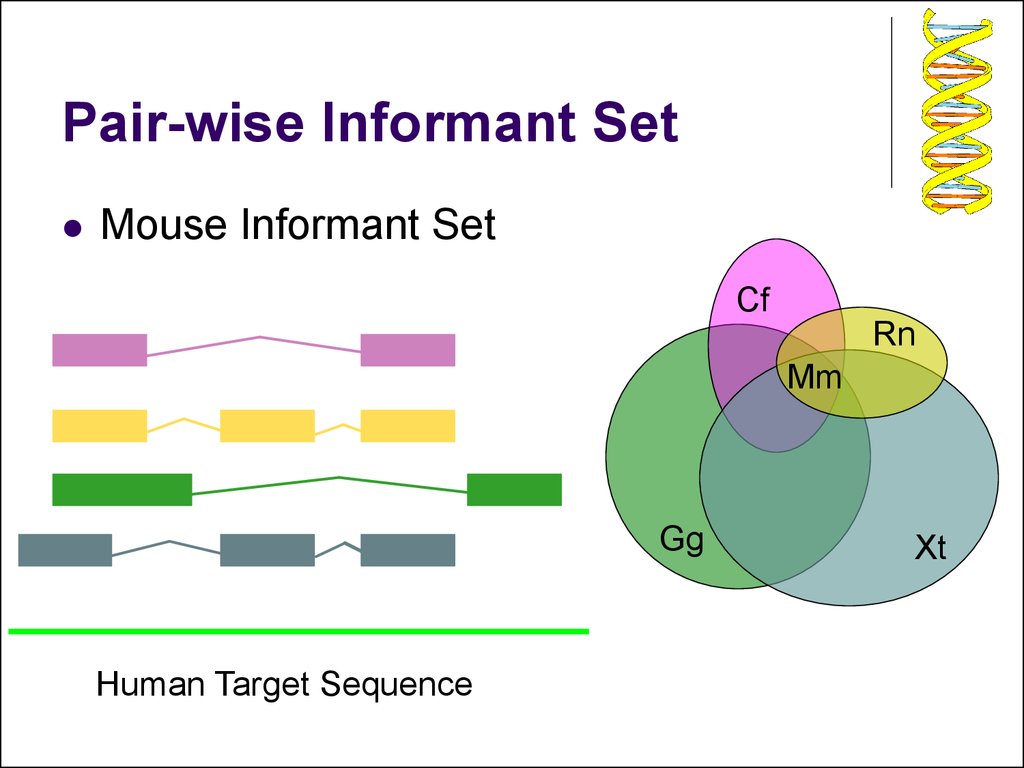
4. Pair-wise Informant Set
Mouse Informant SetCf
Rn
Mm
Gg
Human Target Sequence
Xt
5. Results
SuccessesSuccessfully predicts alternatively spliced
transcripts de novo
Scales well
Approximately 1.67 transcripts per gene with the
mouse informant set
Sensitive…
6. Results
ChallengesTranscript clustering in gene dense regions (like
the manually picked ENCODE regions) can be
problematic




 Реклама
Реклама








